Additional examples of WatFinder utility¶
Here, we present several more case studies on how to use WatFinder.
The files needed for these case studies can be found in here.
Case study 1¶
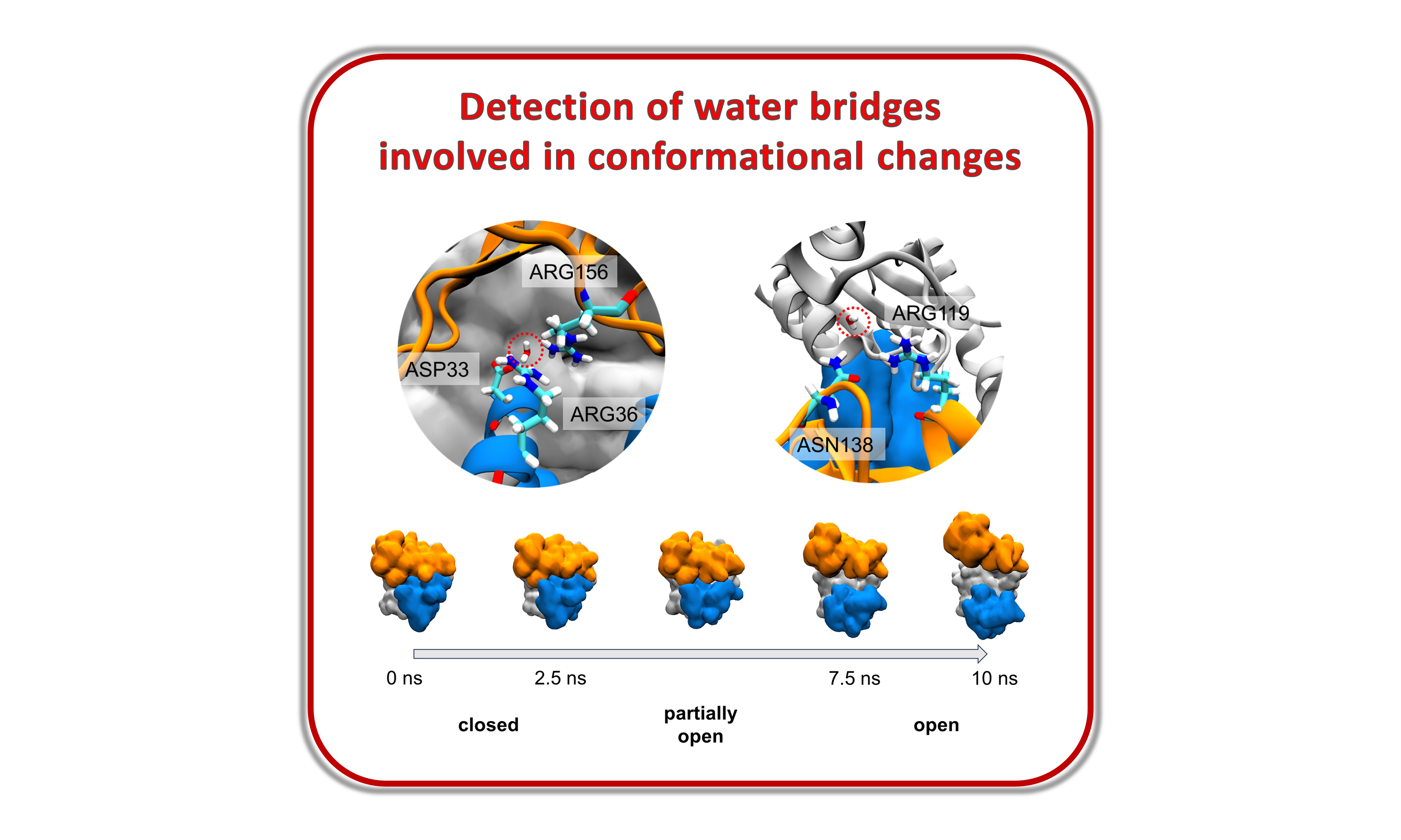
Detection and quantification of the formation of inter-residue water bridges as adenylate kinase transitions from the closed to open state
IPython notebook (ipynb): caseStudy1
Case study 2¶
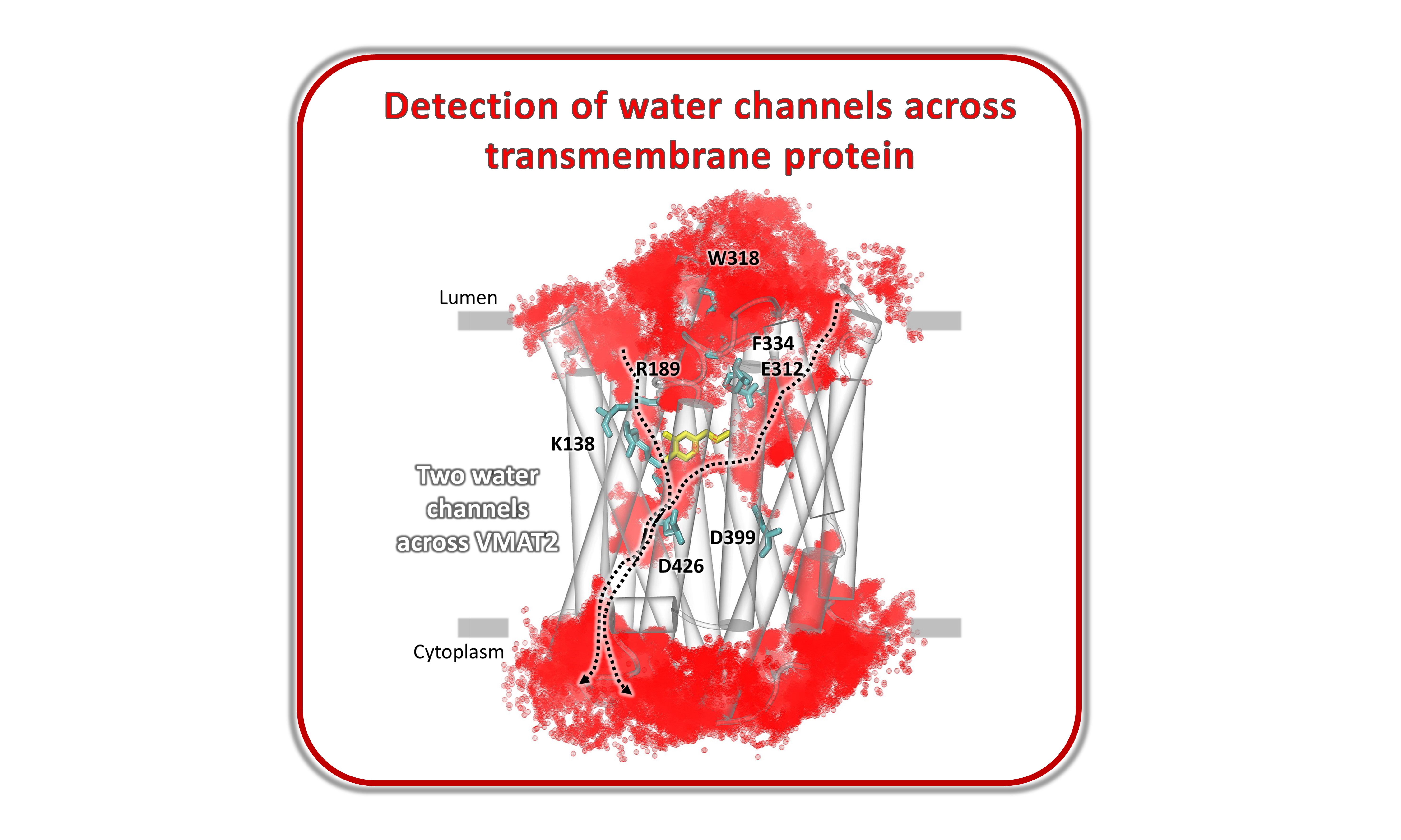
Identification of water influx and clusters into the vesicular monoamine transporter VMAT2
IPython notebook (ipynb): caseStudy2
Case study 3¶
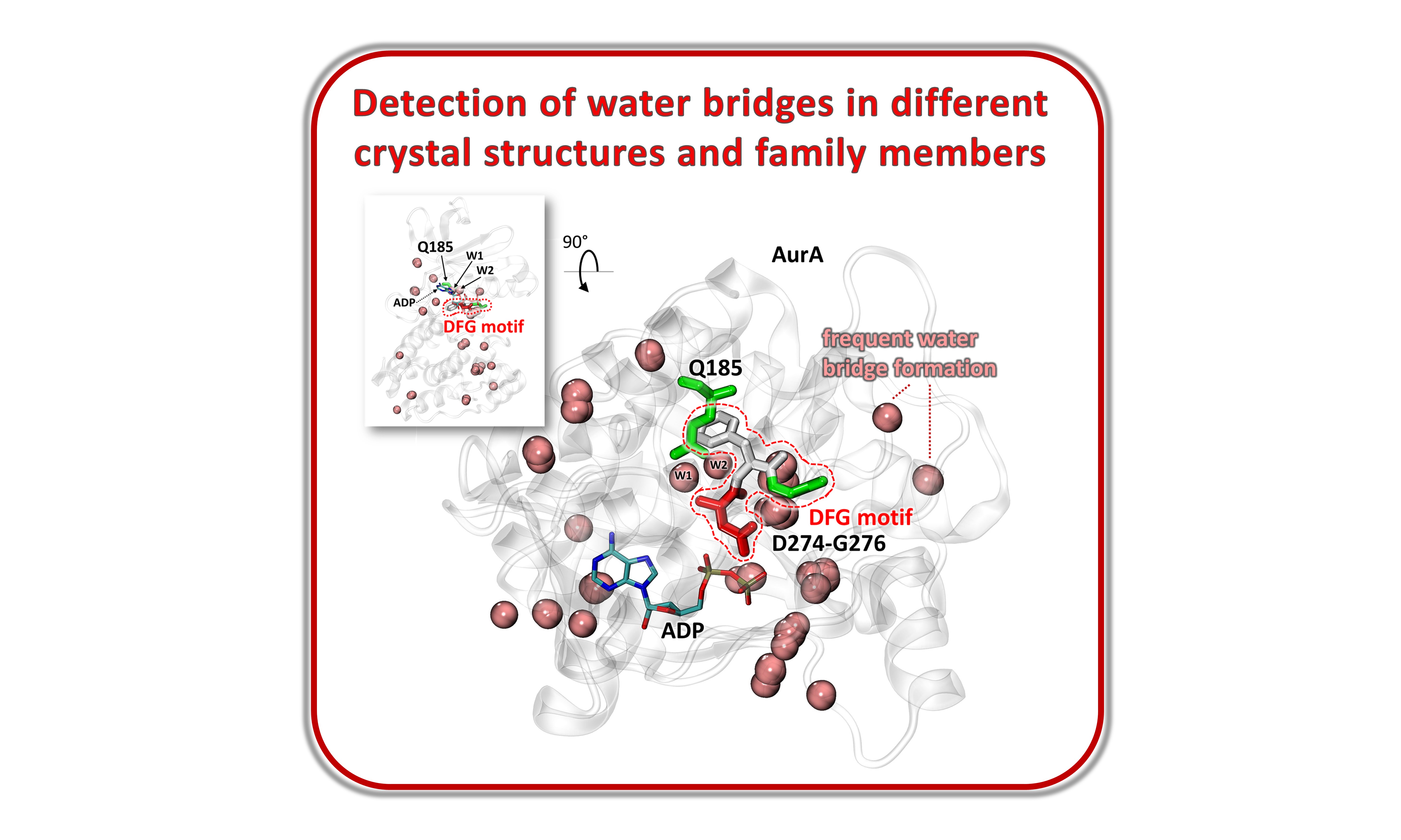
Identification of key protein-water interactions observed across various Aurora kinase A crystal structures identified by BLAST
IPython notebook (ipynb): caseStudy3
Case study 4¶
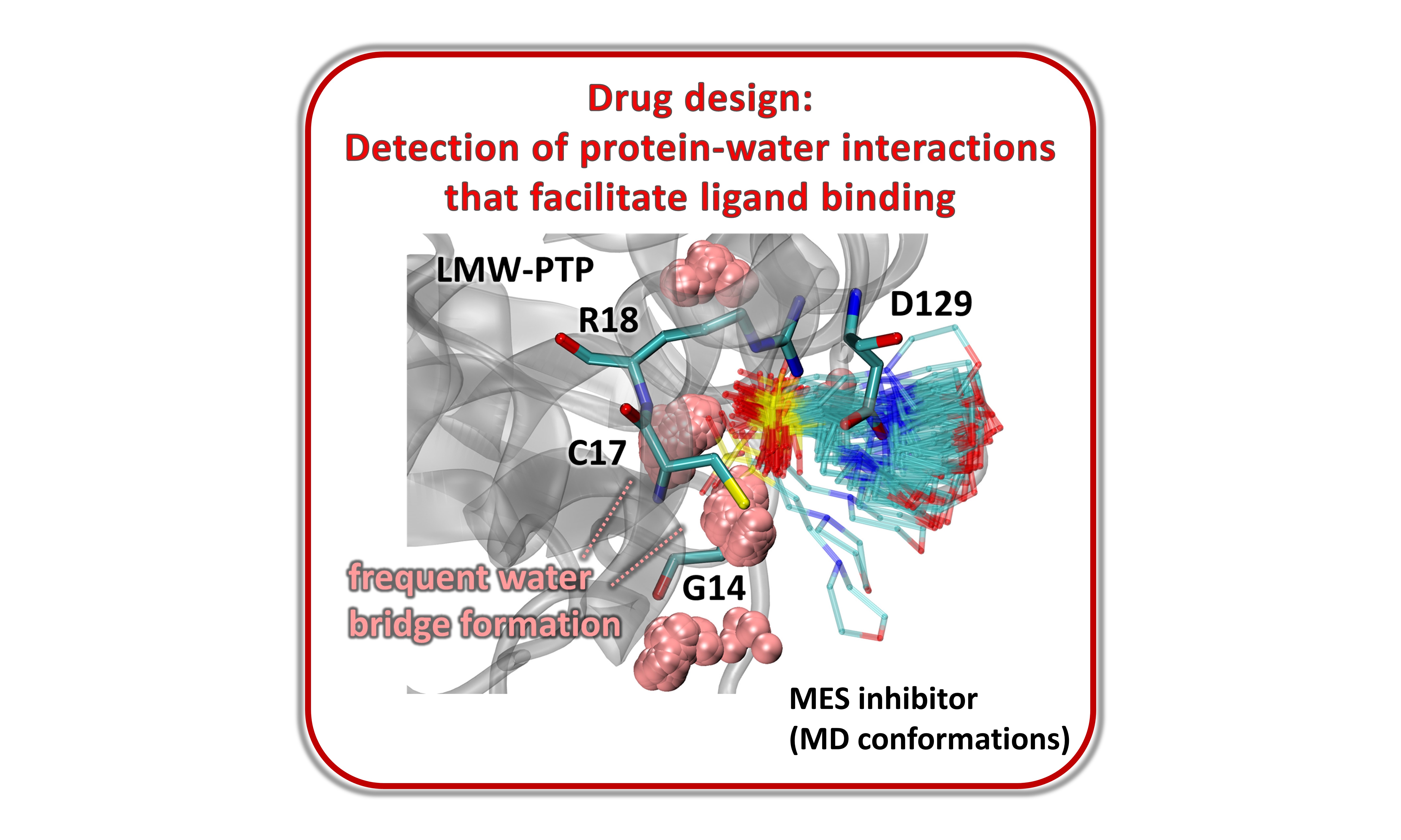
Derection of protein-water interactions that facilitate ligand binding
IPython notebooks (ipynb): caseStudy4a (protein only) and caseStudy4b (protein+ligand)
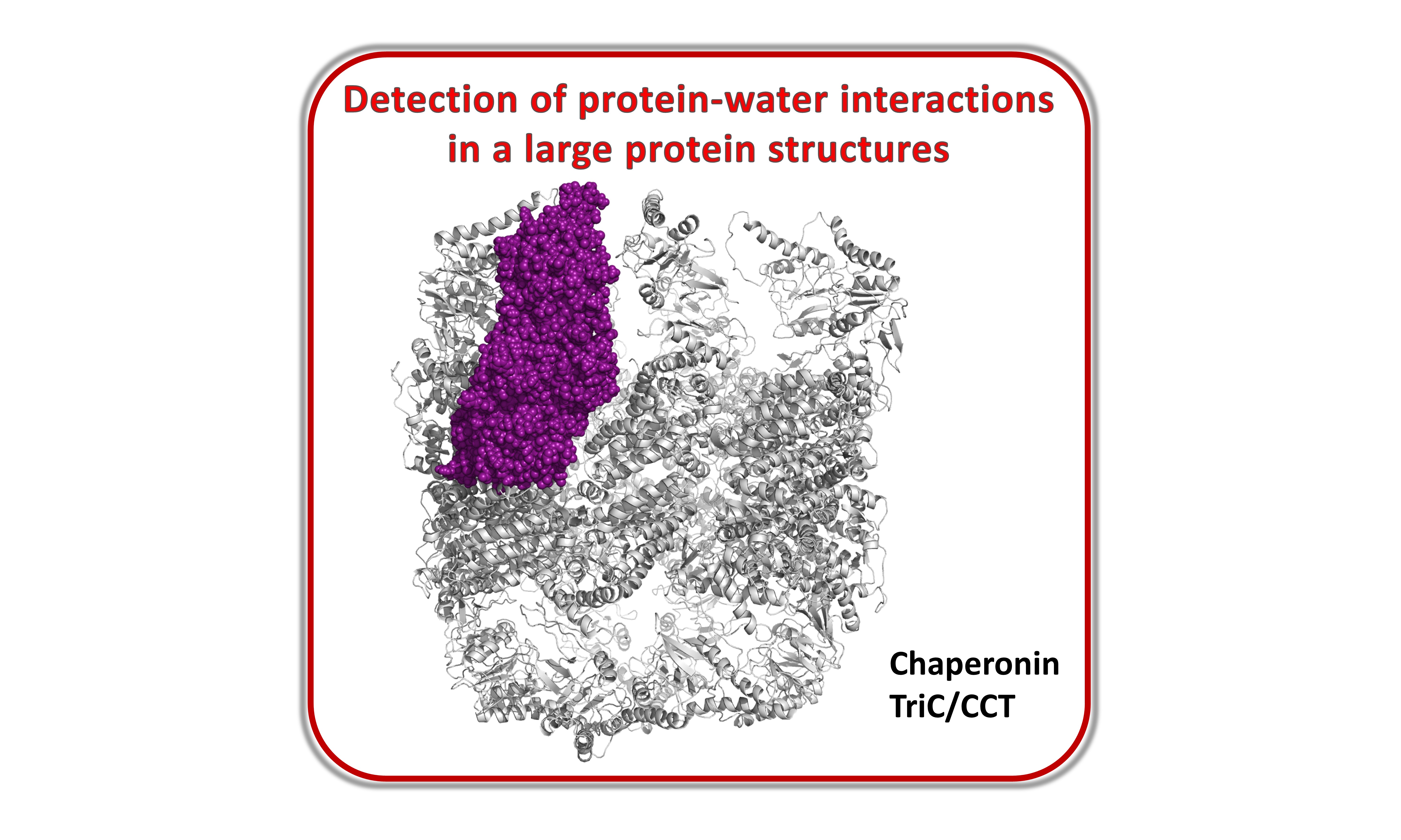
Case study 5¶
Water bridges detection in a large chaperonin TRiC/CCT structure including timing for selected regions
IPython notebook (ipynb): caseStudy5
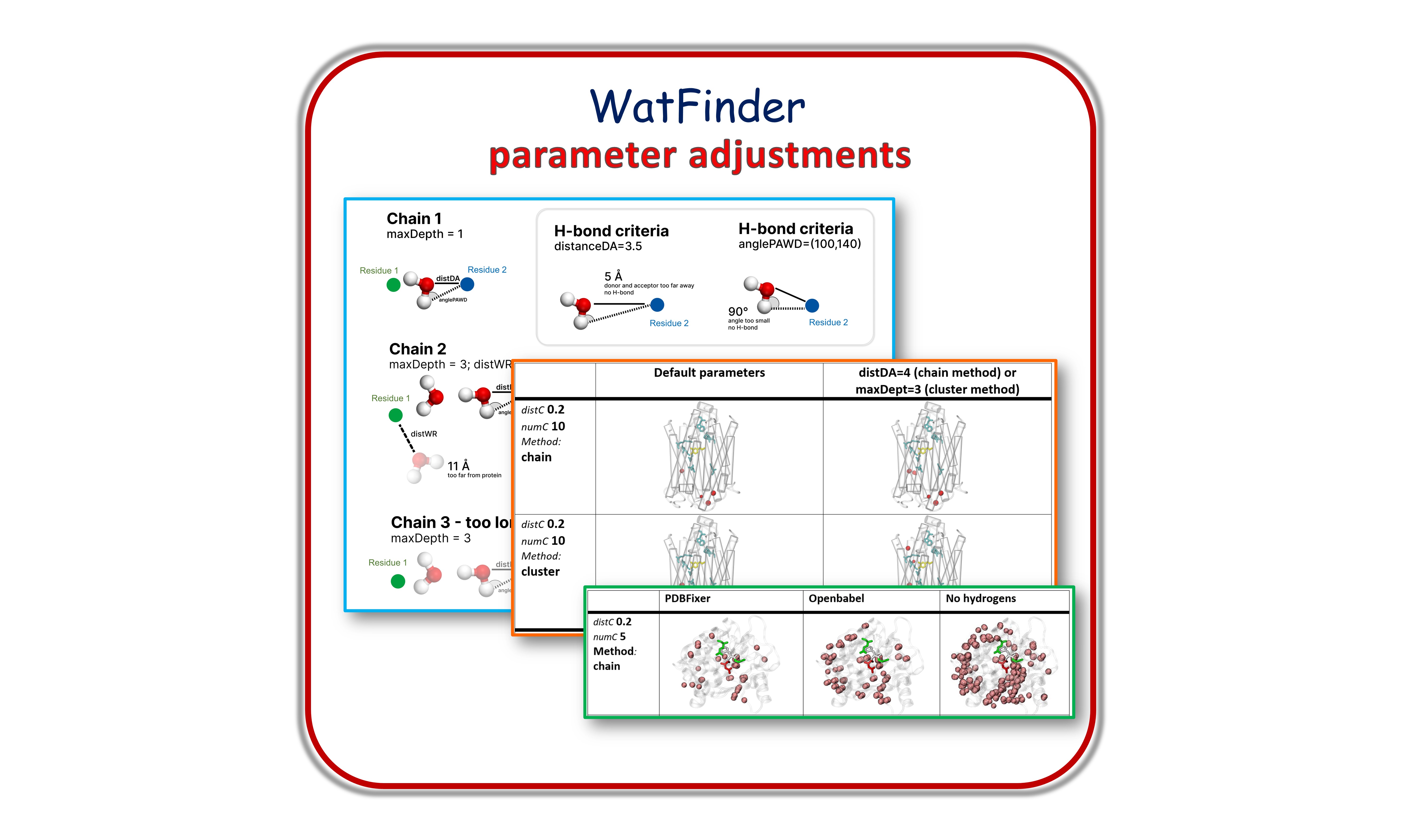
Example 6: WatFinder Parameter Adjustments¶
IPython notebook (ipynb): watfinder_tab1_1
IPython notebook (ipynb): watfinder_tab1_2
IPython notebook (ipynb): watfinder_tab1_3
IPython notebook (ipynb): watfinder_tab1_4
IPython notebook (ipynb): watfinder_tab1_5
IPython notebook (ipynb): watfinder_tab2_1
IPython notebook (ipynb): watfinder_tab2_2
IPython notebook (ipynb): watfinder_tab2_3
IPython notebook (ipynb): watfinder_tab2_4
The description of all cases are available in the supplementary file of the WatFinder paper [JK24]. Trajectories are in here.