Interactions/Stability Evaluation¶
This example shows how to perform Interactions/Stability Evaluation (InSty) analysis for a small protein (<200 residues) called tyrosine phosphatase LMW-PTP (PDB: 5KQM) and visualize the results using Matplotlib library and VMD program. In the tutorial, we will use an already prepared structure for simulation (with hydrogens added). The same structure will be later analyzed with the trajectory file to show how the analysis of interactions in the course of the simulation can change. The file is available in tutorial files.
The tutorial will also include an example of a PDB structure directly downloaded from Protein Data Bank (PDB) which requires adding the missing hydrogen atoms to the protein and ligand structure.
Analysis of interactions for a single PDB structure¶
We start by parsing PDB file with LMW-PTP 5kqm_all_sci.pdb which is avalable
in the tutorial files. The PDB file contains protein structure with water and
counter ions prepared using VMD program.
Before that import everything from the ProDy packages.
In [1]: from prody import *
In [2]: from pylab import *
In [3]: import matplotlib
In [4]: PDBfile = '5kqm_all_sci.pdb'
In [5]: coords = parsePDB(PDBfile)
In [6]: coords
@> 19321 atoms and 1 coordinate set(s) were parsed in 0.23s.
For the analysis we will use only protein coordinates (atoms):
In [7]: atoms = coords.select('protein')
In [8]: atoms
@> 19321 atoms and 1 coordinate set(s) were parsed in 0.21s.
Compute all types of interactions¶
In the next step, we instantiate an Interactions instance:
In [9]: interactions = Interactions()
Now we can compute all available types of interactions (seven types: hydrogen
bonds, salt bridges, repulsive ionic bonding, Pi-cation, Pi-stacking,
hydrophobic interactions, and disulfide bonds) for protein structure by passing
selected atoms (atoms) to Interactions.calcProteinInteractions() method:
In [10]: all_interactions = interactions.calcProteinInteractions(atoms)
@> Calculating interations.
@> Calculating hydrogen bonds.
@> DONOR (res chid atom) <---> ACCEPTOR (res chid atom) Distance Angle
@> ARG101 P NH1_1516 <---> ASP98 P OD1_1463 2.0 33.1
@> HSE72 P NE2_1042 <---> ASN15 P OD1_165 2.6 34.8
@> GLN143 P NE2_2192 <---> GLU139 P OE2_2126 2.7 9.2
@> HSE66 P NE2_957 <---> GLU139 P OE1_2125 2.7 6.4
@> ARG40 P N_561 <---> LYS6 P O_37 2.7 17.1
@> ARG58 P N_813 <---> ASP56 P OD1_788 2.7 30.0
@> ALA45 P N_634 <---> ARG75 P O_1097 2.8 35.1
@> ASN53 P ND2_747 <---> GLU50 P OE1_708 2.8 18.2
@> ALA74 P N_1064 <---> ASN53 P O_751 2.8 21.3
@> ASP56 P N_780 <---> ILE16 P O_189 2.8 27.0
@> LYS110 P NZ_1667 <---> THR84 P O_1240 2.8 38.2
@> LEU116 P N_1758 <---> CYS90 P O_1342 2.8 15.0
@> SER103 P N_1546 <---> LEU99 P O_1485 2.8 29.1
@> ASN134 P N_2045 <---> ASP137 P OD2_2091 2.8 22.6
@> PHE152 P N_2321 <---> CYS148 P O_2275 2.8 8.3
@> ASN95 P N_1398 <---> ASP92 P OD1_1368 2.8 12.6
@> LYS6 P N_16 <---> ASN38 P O_536 2.8 25.0
@> ILE77 P N_1115 <---> ALA45 P O_643 2.8 12.2
@> ARG58 P NH2_832 <---> ASP56 P OD2_789 2.8 27.7
@> LEU99 P N_1467 <---> ASN95 P O_1411 2.8 15.5
@> CYS149 P N_2276 <---> CYS145 P O_2224 2.8 9.6
@> GLY52 P N_731 <---> ALA74 P O_1073 2.8 6.6
@> ASP32 P N_435 <---> LYS28 P O_385 2.8 8.8
@> ILE88 P N_1294 <---> LYS112 P O_1704 2.8 17.7
@> GLN143 P N_2180 <---> GLU139 P O_2128 2.8 21.7
@> ARG27 P N_340 <---> GLU23 P O_293 2.8 15.4
@> TYR142 P N_2159 <---> PHE138 P O_2113 2.9 14.2
@> GLY133 P N_2038 <---> PRO130 P O_1995 2.9 25.4
@> PHE26 P N_320 <---> ALA22 P O_278 2.9 4.9
@> ASN15 P ND2_166 <---> SER19 P OG_232 2.9 32.1
@> ARG75 P NH1_1090 <---> ASP81 P OD2_1194 2.9 19.7
@> ARG75 P NH2_1093 <---> ASP42 P OD2_610 2.9 23.5
@> ARG97 P N_1431 <---> GLU93 P O_1386 2.9 22.2
@> ARG65 P NH2_941 <---> GLU139 P OE1_2125 2.9 32.3
@> VAL25 P N_304 <---> ILE21 P O_268 2.9 8.2
@> LEU153 P N_2341 <---> CYS149 P O_2286 2.9 12.5
@> SER7 P N_38 <---> ASP86 P OD2_1270 2.9 39.9
@> ASP86 P N_1261 <---> SER7 P OG_45 2.9 34.7
@> ARG58 P NH2_832 <---> TYR131 P O_2016 2.9 33.1
@> THR46 P N_644 <---> CYS12 P O_130 2.9 36.1
@> GLN144 P N_2197 <---> THR140 P O_2142 2.9 23.3
@> THR78 P N_1134 <---> ASP81 P OD2_1194 2.9 12.4
@> LEU89 P N_1313 <---> LEU9 P O_83 2.9 29.5
@> THR31 P N_421 <---> ARG27 P O_363 2.9 24.1
@> CYS90 P N_1332 <---> GLU114 P O_1738 2.9 24.6
..
..
@> Number of detected hydrogen bonds: 124.
@> Calculating salt bridges.
@> HSE66 P NE2_957 <---> GLU139 P OE1_2125_2126 2.8
@> ASP81 P OD1_1193_1194 <---> ARG75 P NH1_1090_1093 2.9
@> ASP32 P OD1_443_444 <---> LYS28 P NZ_380 3.0
@> ARG101 P NH1_1516_1519 <---> ASP98 P OD1_1463_1464 3.1
@> ARG27 P NH1_356_359 <---> GLU23 P OE1_290_291 3.7
@> GLU139 P OE1_2125_2126 <---> ARG65 P NH1_938_941 3.8
@> LYS102 P NZ_1540 <---> ASP98 P OD1_1463_1464 3.9
@> ARG58 P NH1_829_832 <---> ASP56 P OD1_788_789 3.9
@> ARG18 P NH1_217_220 <---> ASP92 P OD1_1368_1369 4.1
@> GLU114 P OE1_1735_1736 <---> LYS112 P NZ_1699 4.1
@> ASP120 P OD1_1824_1825 <---> ARG147 P NH1_2257_2260 4.2
@> LYS110 P NZ_1667 <---> ASP86 P OD1_1269_1270 4.2
@> GLU114 P OE1_1735_1736 <---> HSE157 P NE2_2418 4.4
@> ARG18 P NH1_217_220 <---> ASP129 P OD1_1978_1979 4.6
@> ARG75 P NH1_1090_1093 <---> ASP42 P OD1_609_610 4.6
@> GLU23 P OE1_290_291 <---> HSE72 P NE2_1042 5.0
@> Number of detected salt bridges: 16.
@> Calculating repulsive ionic bonding.
@> ARG101 P NH1_1516_1519 <---> LYS102 P NZ_1540 4.3
@> Number of detected Repulsive Ionic Bonding interactions: 1.
@> Calculating Pi stacking interactions.
@> HSE66 P 953_954_955_957_959 <---> TYR142 P 2166_2167_2169_2171_2174_2176 3.9 162.1
@> HSE157 P 2414_2415_2416_2418_2420_2423_2424 <---> TYR119 P 1802_1803_1805_1807_1810_1812 4.4 3.0
@> TRP39 P 549_550_551_553_555_557 <---> PHE26 P 327_328_330_332_334_336 4.8 75.5
@> TYR131 P 2003_2004_2006_2008_2011_2013 <---> TYR132 P 2024_2025_2027_2029_2032_2034 4.9 91.4
@> Number of detected Pi stacking interactions: 4.
@> Calculating cation-Pi interactions.
@> PHE85 P 1248_1249_1251_1253_1255_1257 <---> ARG40 P NH1_577_580 3.7
@> HSE66 P 953_954_955_957_959 <---> ARG65 P NH1_938_941 4.5
@> HSE157 P2414_2415_2416_2418_2420_2423_2424 <---> LYS112 P NZ_1699 4.8
@> Number of detected cation-pi interactions: 3.
@> Hydrophobic Overlaping Areas are computed.
@> Calculating hydrophobic interactions.
@> TYR87 P OH_128614s <---> ALA156 P CB_2401 3.0 22.0
@> MET63 P CE_89414s <---> ALA24 P CB_298 3.3 5.2
@> ILE68 P CG2_97614s <---> MET63 P CE_894 3.3 52.4
@> TYR142 P CZ_217114s <---> VAL146 P CG2_2235 3.5 49.7
@> PHE10 P CD1_9214s <---> ALA22 P CB_273 3.5 31.2
@> LYS6 P CD_2614s <---> TRP39 P CZ2_555 3.5 68.7
@> VAL30 P CG1_41114s <---> PHE26 P CE2_336 3.6 21.1
@> ALA111 P CB_167714s <---> ILE88 P CD_1307 3.6 21.2
@> VAL11 P CG2_11414s <---> ILE88 P CG2_1300 3.6 9.3
@> VAL41 P CG2_59514s <---> PHE26 P CD2_334 3.6 16.6
@> PHE152 P CE1_233114s <---> ALA156 P CB_2401 3.7 17.5
@> VAL106 P CG2_159814s <---> LYS79 P CG_1155 3.7 25.1
@> ILE77 P CD_112814s <---> LEU99 P CD2_1480 3.7 12.0
@> PHE82 P CD1_120514s <---> ILE88 P CD_1307 3.7 17.6
@> LEU116 P CD2_177114s <---> ILE127 P CD_1949 3.7 17.4
@> VAL8 P CG1_5514s <---> PHE26 P CE2_336 3.7 12.1
@> LEU96 P CD1_142114s <---> ILE113 P CG2_1711 3.7 17.0
@> LEU9 P CD2_7814s <---> ILE77 P CD_1128 3.7 15.4
@> LEU89 P CD1_132214s <---> VAL8 P CG2_59 3.8 15.9
@> ILE126 P CD_193014s <---> LEU125 P CD1_1907 3.8 54.2
@> VAL141 P CG1_214914s <---> ILE127 P CG2_1942 3.9 11.5
..
..
@> Number of detected hydrophobic interactions: 39.
@> Calculating disulfide bonds.
@> Number of detected disulfide bonds: 0.
All types of interactions will be displayed on the screen with all types of information such as distance or angle (if applied).
Moreover, we will have access to the details of each interaction type using the following methods:
Interactions.getHydrogenBonds() - hydrogen bonds:
In [11]: interactions.getHydrogenBonds()
[['ARG101', 'NH1_1516', 'P', 'ASP98', 'OD1_1463', 'P', 1.998, 33.1238],
['HSE72', 'NE2_1042', 'P', 'ASN15', 'OD1_165', 'P', 2.5997, 34.752],
['GLN143', 'NE2_2192', 'P', 'GLU139', 'OE2_2126', 'P', 2.7287, 9.1823],
['HSE66', 'NE2_957', 'P', 'GLU139', 'OE1_2125', 'P', 2.7314, 6.3592],
['ARG40', 'N_561', 'P', 'LYS6', 'O_37', 'P', 2.7479, 17.1499],
['ARG58', 'N_813', 'P', 'ASP56', 'OD1_788', 'P', 2.7499, 29.9737],
['ALA45', 'N_634', 'P', 'ARG75', 'O_1097', 'P', 2.7609, 35.0983],
['ASN53', 'ND2_747', 'P', 'GLU50', 'OE1_708', 'P', 2.7702, 18.2336],
['ALA74', 'N_1064', 'P', 'ASN53', 'O_751', 'P', 2.7782, 21.3375],
['ASP56', 'N_780', 'P', 'ILE16', 'O_189', 'P', 2.7793, 27.0481],
['LYS110', 'NZ_1667', 'P', 'THR84', 'O_1240', 'P', 2.7977, 38.2213],
['LEU116', 'N_1758', 'P', 'CYS90', 'O_1342', 'P', 2.8072, 15.0239],
['SER103', 'N_1546', 'P', 'LEU99', 'O_1485', 'P', 2.8075, 29.107],
['ASN134', 'N_2045', 'P', 'ASP137', 'OD2_2091', 'P', 2.8132, 22.562],
['PHE152', 'N_2321', 'P', 'CYS148', 'O_2275', 'P', 2.8141, 8.2562],
['ASN95', 'N_1398', 'P', 'ASP92', 'OD1_1368', 'P', 2.8148, 12.5701],
['LYS6', 'N_16', 'P', 'ASN38', 'O_536', 'P', 2.8178, 25.0305],
['ILE77', 'N_1115', 'P', 'ALA45', 'O_643', 'P', 2.8179, 12.1855],
..
..
Interactions.getSaltBridges() - salt bridges (residues with oposite
charges):
In [12]: interactions.getSaltBridges()
[['HSE66', 'NE2_957', 'P', 'GLU139', 'OE1_2125_2126', 'P', 2.8359],
['ASP81', 'OD1_1193_1194', 'P', 'ARG75', 'NH1_1090_1093', 'P', 2.9163],
['ASP32', 'OD1_443_444', 'P', 'LYS28', 'NZ_380', 'P', 3.037],
['ARG101', 'NH1_1516_1519', 'P', 'ASP98', 'OD1_1463_1464', 'P', 3.0699],
['ARG27', 'NH1_356_359', 'P', 'GLU23', 'OE1_290_291', 'P', 3.7148],
['GLU139', 'OE1_2125_2126', 'P', 'ARG65', 'NH1_938_941', 'P', 3.7799],
['LYS102', 'NZ_1540', 'P', 'ASP98', 'OD1_1463_1464', 'P', 3.9359],
['ARG58', 'NH1_829_832', 'P', 'ASP56', 'OD1_788_789', 'P', 3.9486],
['ARG18', 'NH1_217_220', 'P', 'ASP92', 'OD1_1368_1369', 'P', 4.0693],
['GLU114', 'OE1_1735_1736', 'P', 'LYS112', 'NZ_1699', 'P', 4.0787],
['ASP120', 'OD1_1824_1825', 'P', 'ARG147', 'NH1_2257_2260', 'P', 4.1543],
['LYS110', 'NZ_1667', 'P', 'ASP86', 'OD1_1269_1270', 'P', 4.1879],
['GLU114', 'OE1_1735_1736', 'P', 'HSE157', 'NE2_2418', 'P', 4.3835],
['ARG18', 'NH1_217_220', 'P', 'ASP129', 'OD1_1978_1979', 'P', 4.5608],
['ARG75', 'NH1_1090_1093', 'P', 'ASP42', 'OD1_609_610', 'P', 4.5612],
['GLU23', 'OE1_290_291', 'P', 'HSE72', 'NE2_1042', 'P', 4.99]]
Interactions.getRepulsiveIonicBonding() - repulsive ionic bonding
(between residues with the same charges):
In [13]: interactions.getRepulsiveIonicBonding()
[['ARG101', 'NH1_1516_1519', 'P', 'LYS102', 'NZ_1540', 'P', 4.2655]]
Interactions.getPiStacking() - Pi-stacking interactions (HSE is a histidine
(HIS) type in the CHARMM force field):
In [14]: interactions.getPiStacking()
[['HSE66',
'953_954_955_957_959',
'P',
'TYR142',
'2166_2167_2169_2171_2174_2176',
'P',
3.8882,
162.1245],
['HSE157',
'2414_2415_2416_2418_2420_2423_2424',
'P',
'TYR119',
'1802_1803_1805_1807_1810_1812',
'P',
4.3605,
3.0062],
['TRP39',
'549_550_551_553_555_557',
'P',
'PHE26',
'327_328_330_332_334_336',
'P',
4.8394,
75.4588],
['TYR131',
'2003_2004_2006_2008_2011_2013',
'P',
'TYR132',
'2024_2025_2027_2029_2032_2034',
'P',
4.8732,
91.4358]]
Interactions.getPiCation() - Pi-cation:
In [15]: interactions.getPiCation()
[['PHE85',
'1248_1249_1251_1253_1255_1257',
'P',
'ARG40',
'NH1_577_580',
'P',
3.6523],
['HSE66', '953_954_955_957_959', 'P', 'ARG65', 'NH1_938_941', 'P', 4.5323],
['HSE157',
'2414_2415_2416_2418_2420_2423_2424',
'P',
'LYS112',
'NZ_1699',
'P',
4.828]]
Interactions.getHydrophobic() - hydrophobic interactions:
In [16]: interactions.getHydrophobic()
[['TYR87', 'OH_1286', 'P', 'ALA156', 'CB_2401', 'P', 3.0459],
['MET63', 'CE_894', 'P', 'ALA24', 'CB_298', 'P', 3.3105],
['ILE68', 'CG2_976', 'P', 'MET63', 'CE_894', 'P', 3.3306],
['TYR142', 'CZ_2171', 'P', 'VAL146', 'CG2_2235', 'P', 3.4815],
['PHE10', 'CD1_92', 'P', 'ALA22', 'CB_273', 'P', 3.5334],
['LYS6', 'CD_26', 'P', 'TRP39', 'CZ2_555', 'P', 3.5427],
['VAL30', 'CG1_411', 'P', 'PHE26', 'CE2_336', 'P', 3.5603],
['ALA111', 'CB_1677', 'P', 'ILE88', 'CD_1307', 'P', 3.5627],
['VAL11', 'CG2_114', 'P', 'ILE88', 'CG2_1300', 'P', 3.6386],
['VAL41', 'CG2_595', 'P', 'PHE26', 'CD2_334', 'P', 3.6448],
['PHE152', 'CE1_2331', 'P', 'ALA156', 'CB_2401', 'P', 3.6594],
['VAL106', 'CG2_1598', 'P', 'LYS79', 'CG_1155', 'P', 3.6828],
['ILE77', 'CD_1128', 'P', 'LEU99', 'CD2_1480', 'P', 3.6917],
['PHE82', 'CD1_1205', 'P', 'ILE88', 'CD_1307', 'P', 3.692],
['LEU116', 'CD2_1771', 'P', 'ILE127', 'CD_1949', 'P', 3.7057],
['VAL8', 'CG1_55', 'P', 'PHE26', 'CE2_336', 'P', 3.7106],
..
..
Interactions.getDisulfideBonds() - disulfide bonds (none in the
structure):
In [17]: interactions.getDisulfideBonds()
[]
To display residues with the biggest number of potential interactions and their
types, we can use Interactions.getFrequentInteractors() method:
In [18]: interactions.getFrequentInteractors(contacts_min=4)
@> VAL8P <---> hb:ASP42P hp:LEU89P hp:PHE26P hb:ARG40P
@> LEU9P <---> hp:ALA44P hp:PHE85P hb:LEU89P hp:ILE77P hb:TYR87P
@> CYS12P <---> hb:ASN15P hb:SER19P hb:THR46P hb:ALA44P
@> ASN15P <---> hb:HIS72P hb:ARG75P hb:CYS12P hb:SER19P hb:SER43P
@> ARG18P <---> hb:ALA22P hb:ASP92P sb:ASP92P hb:ILE127P sb:ASP129P hp:VAL141P
@> GLU23P <---> hb:ARG27P hb:ARG27P hb:ARG27P sb:ARG27P sb:HIS72P hb:SER19P
@> VAL25P <---> hb:LEU29P hp:LEU29P hb:ILE21P hp:TYR142P
@> PHE26P <---> hp:VAL8P hb:VAL30P hp:VAL30P ps:TRP39P hp:VAL41P hb:ALA22P
@> ARG27P <---> hb:THR31P hb:GLU23P hb:GLU23P hb:GLU23P sb:GLU23P
@> LYS28P <---> hb:ASP32P sb:ASP32P hb:ALA24P hp:ILE68P
@> TRP39P <---> hp:LYS6P hp:ILE35P hp:LEU153P ps:PHE26P hb:SER36P
@> ARG40P <---> hb:VAL8P pc:PHE85P hb:LYS6P hb:THR84P hp:PHE85P
@> ASP42P <---> hb:PHE10P hb:ARG75P sb:ARG75P hb:VAL8P
@> ALA44P <---> hb:CYS12P hp:ARG75P hp:LEU9P hb:PHE10P
@> ASP56P <---> hb:ARG58P hb:ARG58P hb:ARG58P sb:ARG58P hb:GLN60P hb:ILE16P
@> ARG58P <---> hb:CYS62P hb:ASP56P hb:ASP56P hb:ASP56P sb:ASP56P hb:TYR131P hb:TYR131P hp:PHE138P
@> MET63P <---> hp:ILE21P hb:ILE68P hp:ILE68P hp:ALA24P hb:GLY59P
@> ARG65P <---> pc:HIS66P sb:GLU139P hb:SER61P hb:GLU139P
@> HIS66P <---> ps:TYR142P pc:ARG65P hb:GLU139P sb:GLU139P
@> ARG75P <---> hb:ALA45P sb:ASP81P hb:ASN15P hb:ASP42P sb:ASP42P hp:ALA44P hb:ASP81P hb:ASP81P
@> ILE77P <---> hp:LEU9P hp:LYS102P hb:ALA45P hp:LEU99P
@> ASP81P <---> hb:ARG75P hb:ARG75P hb:THR78P hb:PHE85P sb:ARG75P hb:THR78P
@> PHE85P <---> hp:ARG40P hp:LEU9P pc:ARG40P hb:ASP81P
@> ASP86P <---> hb:SER7P sb:LYS110P hb:LYS112P hb:SER7P
@> ILE88P <---> hp:VAL11P hp:PHE82P hp:ALA111P hb:GLU114P hb:LYS112P
@> LEU89P <---> hb:VAL11P hp:TYR119P hp:VAL8P hb:LEU9P
@> ASP92P <---> hb:ARG18P sb:ARG18P hb:ASN95P hb:LEU96P
@> ASP98P <---> hb:ARG101P sb:ARG101P hb:LYS102P hb:LYS102P sb:LYS102P hb:SER94P
@> LYS102P <---> rb:ARG101P hb:GLN105P hp:ILE77P hb:ASP98P hb:ASP98P sb:ASP98P
@> LYS112P <---> hb:ILE88P sb:GLU114P pc:HIS157P hb:ASP86P hb:HIS157P
@> GLU114P <---> hb:CYS90P hb:ILE88P sb:LYS112P sb:HIS157P
@> TYR119P <---> hb:HIS157P hp:LEU89P hb:HIS157P ps:HIS157P
@> ASP120P <---> hb:LYS123P hb:GLN124P sb:ARG147P hb:GLY117P
@> ILE127P <---> hb:ARG18P hp:MET91P hp:LEU116P hp:VAL141P hb:MET91P
@> TYR131P <---> hb:ARG58P hb:ARG58P ps:TYR132P hp:ILE16P
@> PHE138P <---> hp:ARG58P hb:TYR142P hp:ILE21P hb:ASN134P
@> GLU139P <---> hb:ARG65P hb:HIS66P sb:HIS66P hb:GLN143P hb:GLN143P sb:ARG65P hb:ASP135P
@> VAL141P <---> hp:ARG18P hb:CYS145P hp:ILE127P hb:ASP137P
@> TYR142P <---> hp:VAL25P hb:VAL146P ps:HIS66P hb:PHE138P hp:VAL146P
@> ARG147P <---> hb:ALA151P sb:ASP120P hb:GLN124P hb:GLN124P hb:GLN143P
@> HIS157P <---> hb:LYS112P sb:GLU114P hb:TYR119P ps:TYR119P pc:LYS112P hb:TYR119P
@> ARG101P <---> hb:ARG97P hb:ASP98P sb:ASP98P rb:LYS102P
@>
Legend: hb-hydrogen bond, sb-salt bridge, rb-repulsive ionic bond, ps-Pi stacking interaction,
pc-Cation-Pi interaction, hp-hydrophobic interaction, dibs-disulfide bonds
The value of contacts_min can be modified to display residues with smaller
or bigger number of interactions.
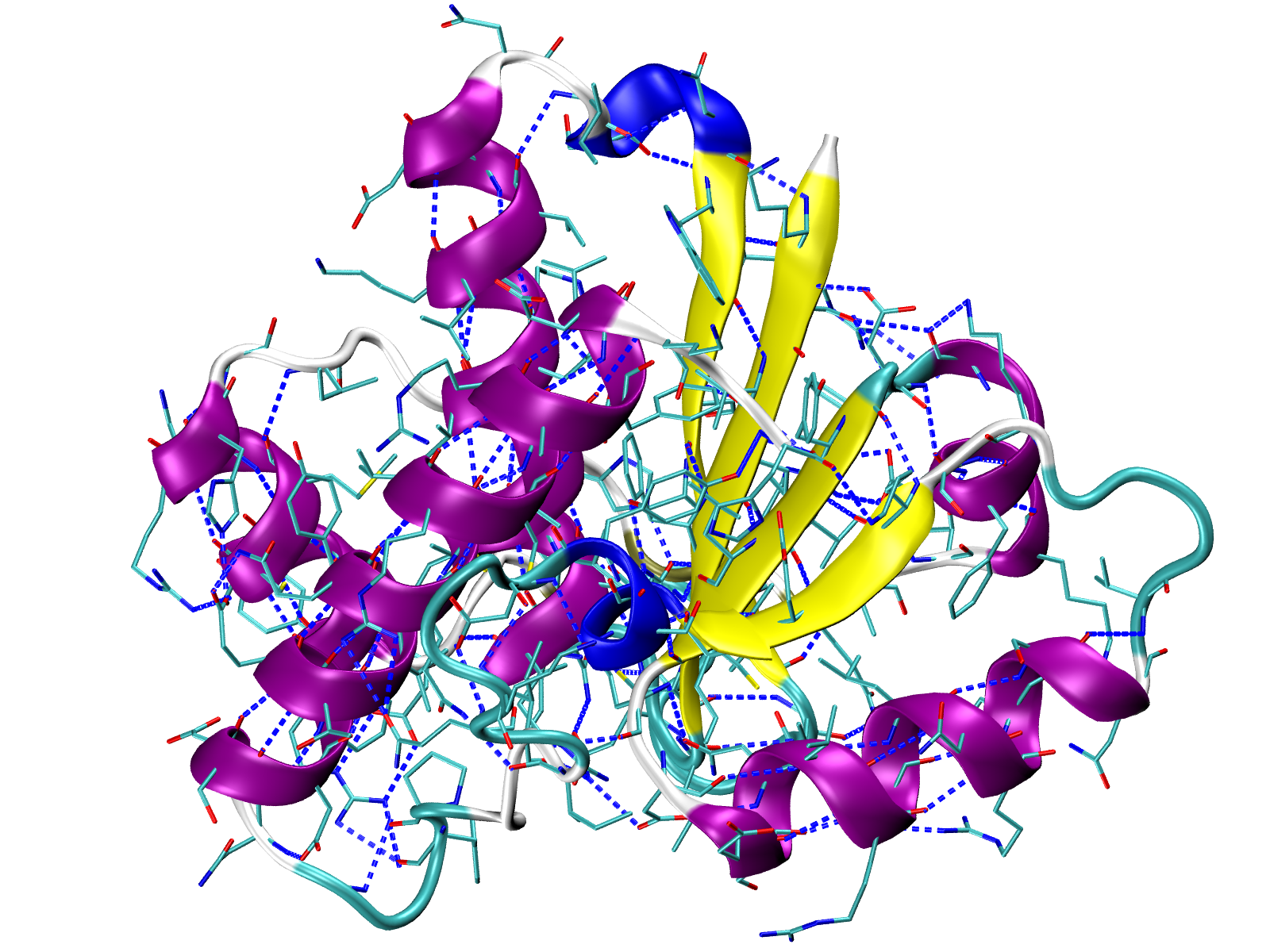
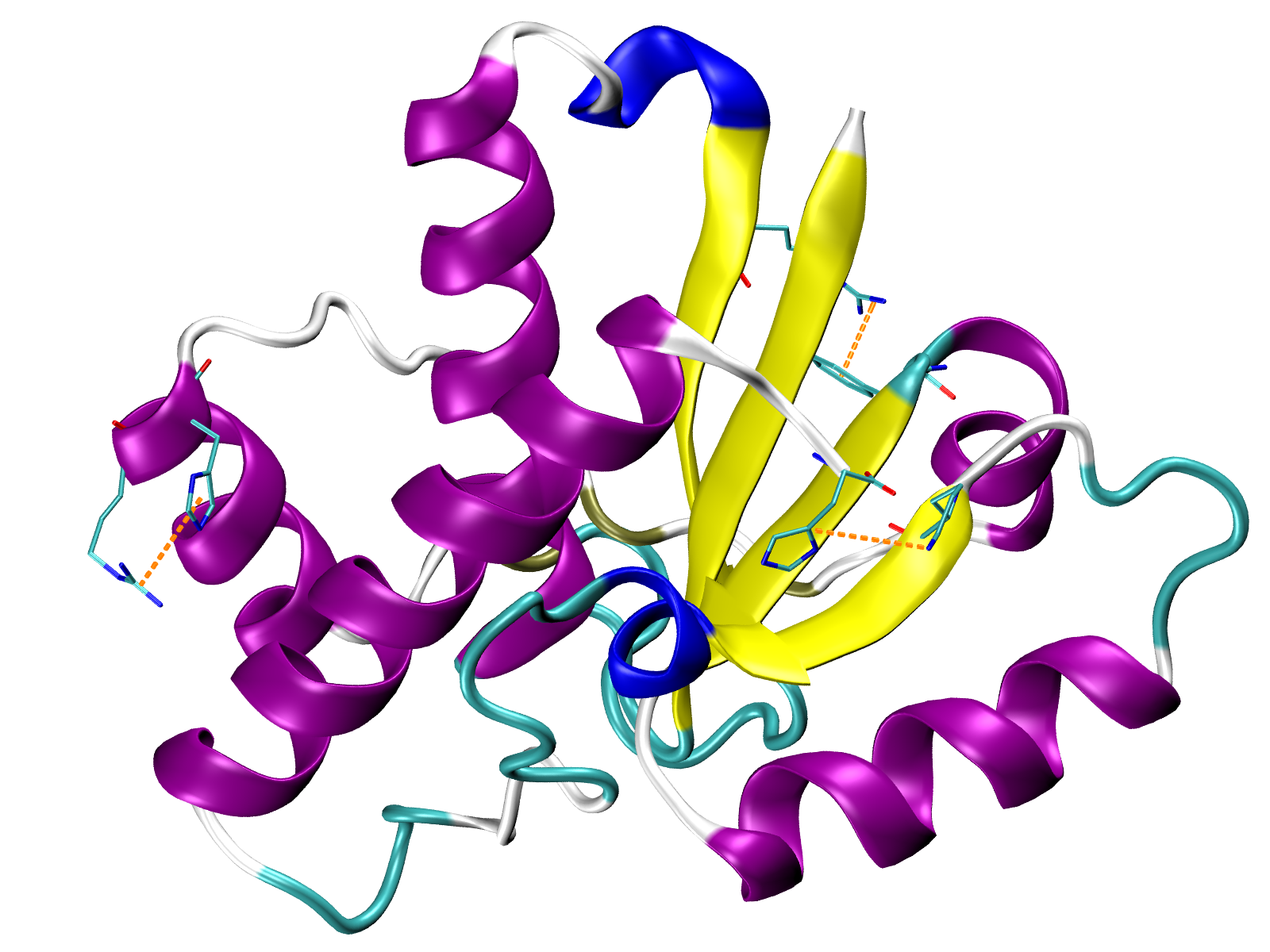
Visualize interactions in VMD¶
We can generate tcl files for visualizing each type of interaction with VMD
using the showProteinInteractions_VMD() function in the following way:
In [19]: showProteinInteractions_VMD(atoms, interactions.getHydrogenBonds(),
....: color='blue', filename='HBs.tcl')
....:
In [20]: showProteinInteractions_VMD(atoms, interactions.getSaltBridges(),
....: color='yellow',filename='SBs.tcl')
....:
In [21]: showProteinInteractions_VMD(atoms, interactions.getRepulsiveIonicBonding(),
....: color='red',filename='RIB.tcl')
....:
In [22]: showProteinInteractions_VMD(atoms, interactions.getPiStacking(),
....: color='green',filename='PiStacking.tcl')
....:
In [23]: showProteinInteractions_VMD(atoms, interactions.getPiCation(),
....: color='orange',filename='PiCation.tcl')
....:
In [24]: showProteinInteractions_VMD(atoms, interactions.getHydrophobic(),
....: color='silver',filename='HPh.tcl')
....:
In [25]: showProteinInteractions_VMD(atoms, interactions.getDisulfideBonds(),
....: color='black',filename='DiBs.tcl')
....:
@> TCL file saved
@> TCL file saved
@> TCL file saved
@> TCL file saved
@> TCL file saved
@> TCL file saved
@> Lack of results
@> TCL file saved
A TCL file will be saved and can be used in VMD after uploading the PDB file
with protein structure 5kqm_all_sci.pdb and by running the following command
line instruction in the VMD TK Console (via VMD Main)
for Linux, Windows and Mac users:
play HBs.tcl
The tcl file contains a method for drawing lines between selected pairs of
residues. Those residues are also displayed. Now, we uploaded hydrogen
bonds which are displayed in blue as we defined in
showProteinInteractions_VMD() function.
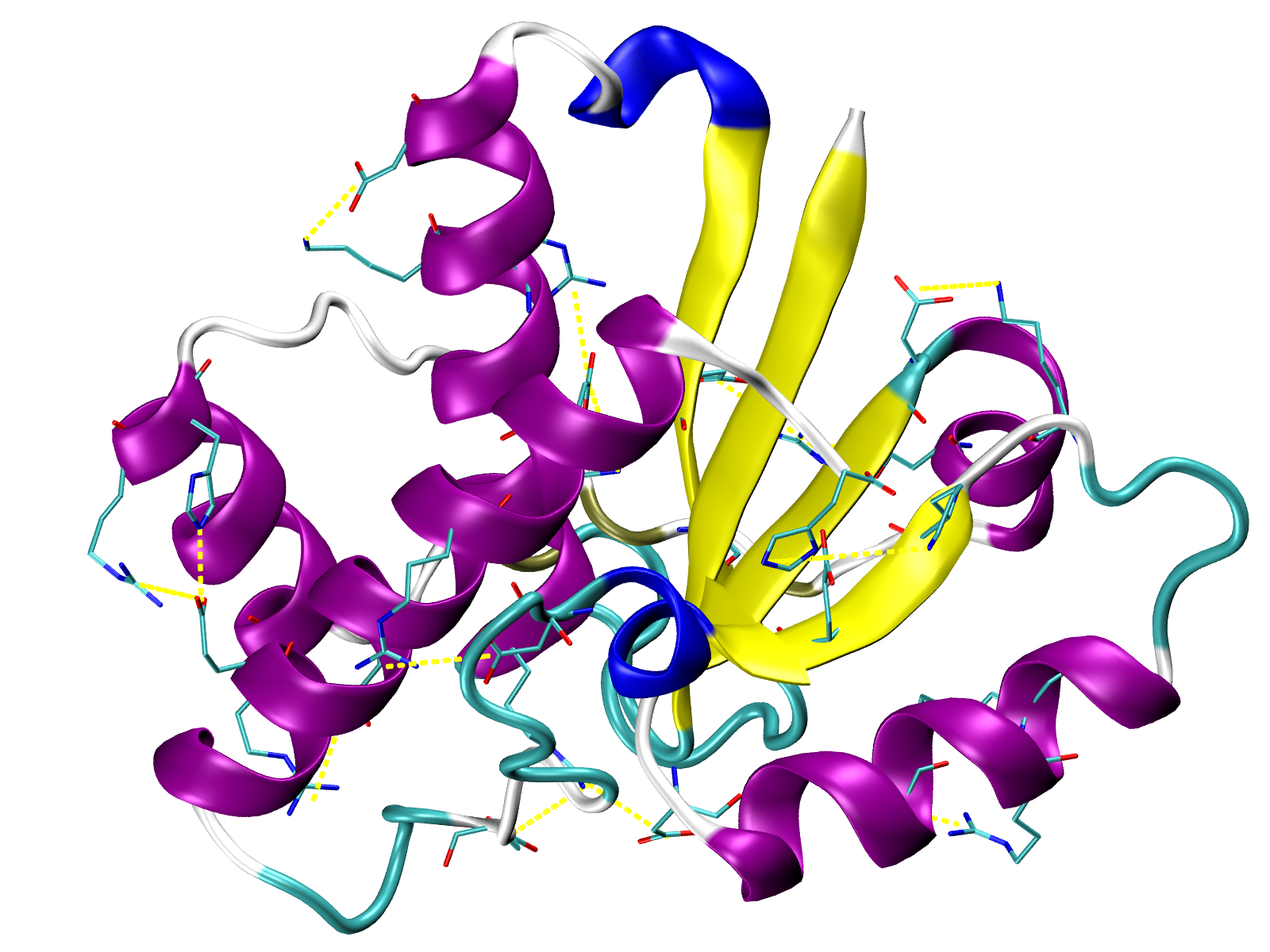
Salt bridges in yellow (VMD TK Console):
play SBs.tcl
Repulsive ionic bonding in red (VMD TK Console):
play RIB.tcl
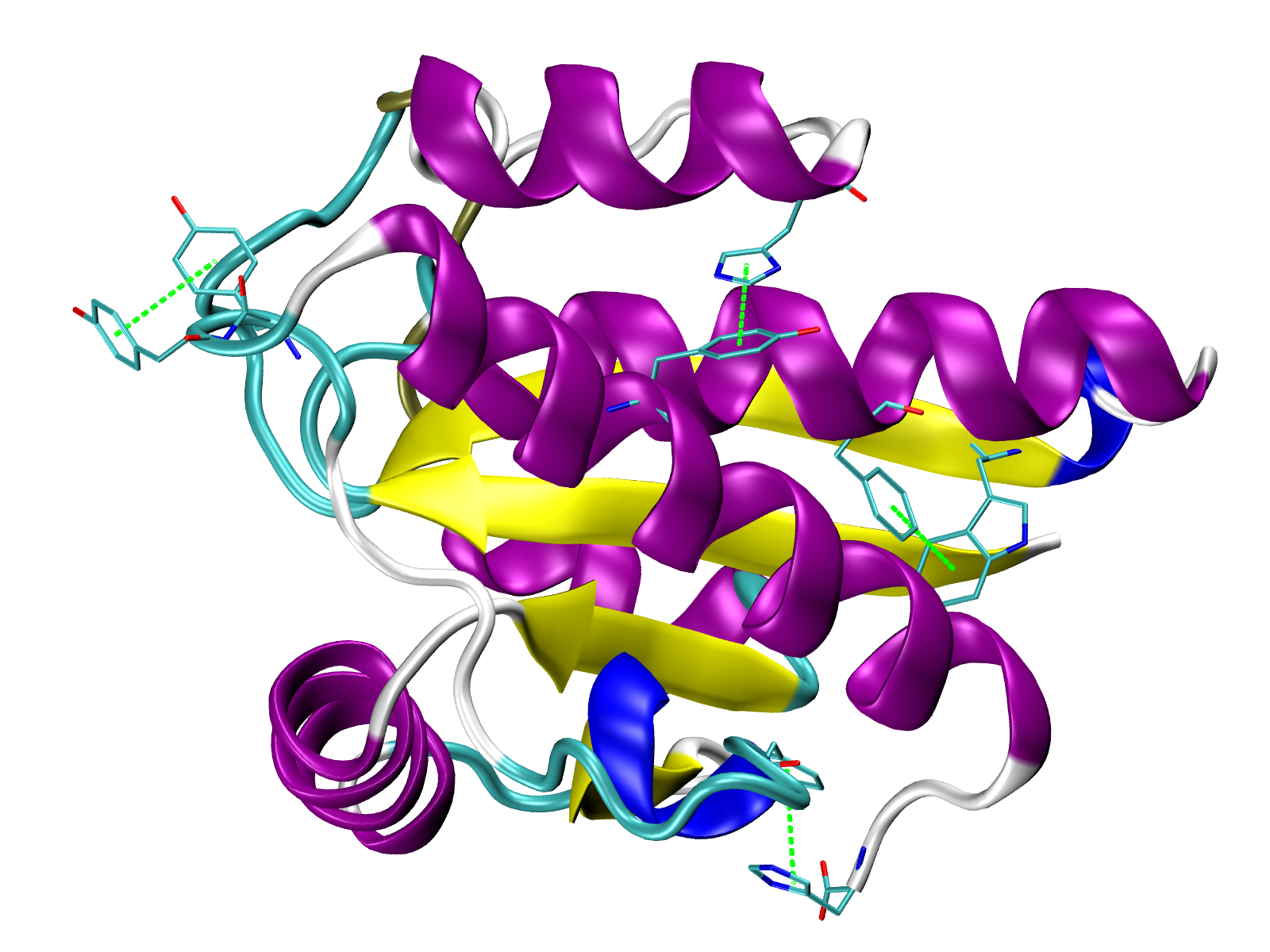
Pi-Pi stacking interactions in green (VMD TK Console):
play PiStacking.tcl
Pi-cation interactions in orange (VMD TK Console):
play PiCation.tcl
and hydrophobic interactions in grey (VMD TK Console):
play HPh.tcl
Additional selections¶
From the predicted interactions, we can select only interactions assigned to certain regions, chains, or between different chains (binding interface between two chains in protein complex).
We can compute them by adding additional parameters to the selected function. See examples below:
In [26]: interactions.getSaltBridges(selection='chain P')
[['HSE66', 'NE2_957', 'P', 'GLU139', 'OE1_2125_2126', 'P', 2.8359],
['ASP81', 'OD1_1193_1194', 'P', 'ARG75', 'NH1_1090_1093', 'P', 2.9163],
['ASP32', 'OD1_443_444', 'P', 'LYS28', 'NZ_380', 'P', 3.037],
['ARG101', 'NH1_1516_1519', 'P', 'ASP98', 'OD1_1463_1464', 'P', 3.0699],
['ARG27', 'NH1_356_359', 'P', 'GLU23', 'OE1_290_291', 'P', 3.7148],
['GLU139', 'OE1_2125_2126', 'P', 'ARG65', 'NH1_938_941', 'P', 3.7799],
['LYS102', 'NZ_1540', 'P', 'ASP98', 'OD1_1463_1464', 'P', 3.9359],
['ARG58', 'NH1_829_832', 'P', 'ASP56', 'OD1_788_789', 'P', 3.9486],
['ARG18', 'NH1_217_220', 'P', 'ASP92', 'OD1_1368_1369', 'P', 4.0693],
['GLU114', 'OE1_1735_1736', 'P', 'LYS112', 'NZ_1699', 'P', 4.0787],
['ASP120', 'OD1_1824_1825', 'P', 'ARG147', 'NH1_2257_2260', 'P', 4.1543],
['LYS110', 'NZ_1667', 'P', 'ASP86', 'OD1_1269_1270', 'P', 4.1879],
['GLU114', 'OE1_1735_1736', 'P', 'HSE157', 'NE2_2418', 'P', 4.3835],
['ARG18', 'NH1_217_220', 'P', 'ASP129', 'OD1_1978_1979', 'P', 4.5608],
['ARG75', 'NH1_1090_1093', 'P', 'ASP42', 'OD1_609_610', 'P', 4.5612],
['GLU23', 'OE1_290_291', 'P', 'HSE72', 'NE2_1042', 'P', 4.99]]
In [27]: interactions.getRepulsiveIonicBonding(selection='resid 102')
[['ARG101', 'NH1_1516_1519', 'P', 'LYS102', 'NZ_1540', 'P', 4.2655]]
In [28]: interactions.getPiStacking(selection='chain P and resid 26')
[['TRP39',
'549_550_551_553_555_557',
'P',
'PHE26',
'327_328_330_332_334_336',
'P',
4.8394,
75.4588]]
It can be done for all kinds of interactions is a similar way. The function will return a list of interactions with following order:
- Hydrogen bonds
- Salt Bridges
- RepulsiveIonicBonding
- Pi stacking interactions
- Pi-cation interactions
- Hydrophobic interactions
- Disulfide bonds
In [29]: allRes_20to50 = interactions.getInteractions(selection='resid 20 to 50')
In [30]: allRes_20to50
[[['ARG40', 'N_561', 'P', 'LYS6', 'O_37', 'P', 2.7479, 17.1499],
['ALA45', 'N_634', 'P', 'ARG75', 'O_1097', 'P', 2.7609, 35.0983],
['ASN53', 'ND2_747', 'P', 'GLU50', 'OE1_708', 'P', 2.7702, 18.2336],
['LYS6', 'N_16', 'P', 'ASN38', 'O_536', 'P', 2.8178, 25.0305],
['ILE77', 'N_1115', 'P', 'ALA45', 'O_643', 'P', 2.8179, 12.1855],
['ASP32', 'N_435', 'P', 'LYS28', 'O_385', 'P', 2.8357, 8.8318],
['ARG27', 'N_340', 'P', 'GLU23', 'O_293', 'P', 2.8446, 15.4167],
['PHE26', 'N_320', 'P', 'ALA22', 'O_278', 'P', 2.8541, 4.8732],
['ARG75', 'NH2_1093', 'P', 'ASP42', 'OD2_610', 'P', 2.8649, 23.5083],
['VAL25', 'N_304', 'P', 'ILE21', 'O_268', 'P', 2.8666, 8.2255],
['THR46', 'N_644', 'P', 'CYS12', 'O_130', 'P', 2.883, 36.1279],
['THR31', 'N_421', 'P', 'ARG27', 'O_363', 'P', 2.896, 24.1287],
['GLU23', 'N_279', 'P', 'SER19', 'O_235', 'P', 2.8979, 15.4146],
['PHE10', 'N_84', 'P', 'ASP42', 'O_612', 'P', 2.9026, 22.751],
['ARG27', 'NH2_359', 'P', 'GLU23', 'OE2_291', 'P', 2.9199, 31.5487],
['ASN38', 'N_523', 'P', 'ILE35', 'O_496', 'P', 2.9255, 29.091],
['GLN76', 'NE2_1110', 'P', 'THR46', 'O_657', 'P', 2.9381, 31.3836],
['ARG40', 'NH1_577', 'P', 'THR84', 'OG1_1233', 'P', 2.9482, 8.3748],
['ALA44', 'N_624', 'P', 'PHE10', 'O_103', 'P', 2.9499, 33.1772],
['VAL8', 'N_49', 'P', 'ARG40', 'O_584', 'P', 2.9631, 25.0079],
['ILE35', 'N_478', 'P', 'VAL30', 'O_420', 'P', 2.9811, 23.5092],
['ASN53', 'N_738', 'P', 'GLU50', 'O_711', 'P', 2.995, 28.587],
['ASN34', 'N_464', 'P', 'THR31', 'O_434', 'P', 3.0041, 18.2465],
['ASN15', 'ND2_166', 'P', 'SER43', 'OG_620', 'P', 3.0129, 25.6996],
['ARG27', 'NH1_356', 'P', 'GLU23', 'OE2_291', 'P', 3.0175, 36.9343],
['LEU29', 'N_386', 'P', 'VAL25', 'O_319', 'P', 3.0299, 19.109],
['SER47', 'N_658', 'P', 'LEU13', 'O_149', 'P', 3.0386, 28.8029],
['VAL30', 'N_405', 'P', 'PHE26', 'O_339', 'P', 3.0394, 17.6883],
['ALA24', 'N_294', 'P', 'PRO20', 'O_249', 'P', 3.0751, 29.9487],
['LYS28', 'N_364', 'P', 'ALA24', 'O_303', 'P', 3.0783, 19.9504],
['ALA22', 'N_269', 'P', 'ARG18', 'O_224', 'P', 3.088, 21.873],
['ASP42', 'N_601', 'P', 'VAL8', 'O_64', 'P', 3.1331, 35.5671],
['TRP39', 'N_537', 'P', 'SER36', 'O_507', 'P', 3.1343, 15.1776],
['CYS12', 'N_120', 'P', 'ALA44', 'O_633', 'P', 3.3349, 36.1006]],
[['ASP32', 'OD1_443_444', 'P', 'LYS28', 'NZ_380', 'P', 3.037],
['ARG27', 'NH1_356_359', 'P', 'GLU23', 'OE1_290_291', 'P', 3.7148],
['ARG75', 'NH1_1090_1093', 'P', 'ASP42', 'OD1_609_610', 'P', 4.5612],
['GLU23', 'OE1_290_291', 'P', 'HSE72', 'NE2_1042', 'P', 4.99]],
[],
[['TRP39',
'549_550_551_553_555_557',
'P',
'PHE26',
'327_328_330_332_334_336',
'P',
4.8394,
75.4588]],
[['PHE85',
'1248_1249_1251_1253_1255_1257',
'P',
'ARG40',
'NH1_577_580',
'P',
3.6523]],
[['MET63', 'CE_894', 'P', 'ALA24', 'CB_298', 'P', 3.3105],
['PHE10', 'CD1_92', 'P', 'ALA22', 'CB_273', 'P', 3.5334],
['LYS6', 'CD_26', 'P', 'TRP39', 'CZ2_555', 'P', 3.5427],
['VAL30', 'CG1_411', 'P', 'PHE26', 'CE2_336', 'P', 3.5603],
['VAL41', 'CG2_595', 'P', 'PHE26', 'CD2_334', 'P', 3.6448],
['VAL8', 'CG1_55', 'P', 'PHE26', 'CE2_336', 'P', 3.7106],
['ALA44', 'CB_628', 'P', 'LEU9', 'CD1_74', 'P', 3.8992],
['VAL25', 'CG2_314', 'P', 'TYR142', 'CE1_2169', 'P', 3.92],
['ILE21', 'CG2_256', 'P', 'MET63', 'SD_893', 'P', 3.9614],
['LEU153', 'CD1_2350', 'P', 'TRP39', 'NE1_547', 'P', 3.967],
['ILE35', 'CD_491', 'P', 'TRP39', 'NE1_547', 'P', 4.0172],
['LEU29', 'CD1_395', 'P', 'VAL25', 'CG1_310', 'P', 4.0642],
['ARG75', 'CG_1081', 'P', 'ALA44', 'CB_628', 'P', 4.0853],
['ARG40', 'CG_568', 'P', 'PHE85', 'CE2_1257', 'P', 4.2669],
['LYS28', 'CG_371', 'P', 'ILE68', 'CD_983', 'P', 4.2707],
['PHE138', 'CD2_2108', 'P', 'ILE21', 'CD_263', 'P', 4.3082]],
[]]
The list of hydrogen bonds, salt bridges and other types of interactions can be displayed as follows:
Hydrogen bonds:
In [31]: allRes_20to50[0]
[['ARG40', 'N_561', 'P', 'LYS6', 'O_37', 'P', 2.7479, 17.1499],
['ALA45', 'N_634', 'P', 'ARG75', 'O_1097', 'P', 2.7609, 35.0983],
['ASN53', 'ND2_747', 'P', 'GLU50', 'OE1_708', 'P', 2.7702, 18.2336],
['LYS6', 'N_16', 'P', 'ASN38', 'O_536', 'P', 2.8178, 25.0305],
['ILE77', 'N_1115', 'P', 'ALA45', 'O_643', 'P', 2.8179, 12.1855],
['ASP32', 'N_435', 'P', 'LYS28', 'O_385', 'P', 2.8357, 8.8318],
['ARG27', 'N_340', 'P', 'GLU23', 'O_293', 'P', 2.8446, 15.4167],
['PHE26', 'N_320', 'P', 'ALA22', 'O_278', 'P', 2.8541, 4.8732],
['ARG75', 'NH2_1093', 'P', 'ASP42', 'OD2_610', 'P', 2.8649, 23.5083],
['VAL25', 'N_304', 'P', 'ILE21', 'O_268', 'P', 2.8666, 8.2255],
['THR46', 'N_644', 'P', 'CYS12', 'O_130', 'P', 2.883, 36.1279],
['THR31', 'N_421', 'P', 'ARG27', 'O_363', 'P', 2.896, 24.1287],
['GLU23', 'N_279', 'P', 'SER19', 'O_235', 'P', 2.8979, 15.4146],
['PHE10', 'N_84', 'P', 'ASP42', 'O_612', 'P', 2.9026, 22.751],
['ARG27', 'NH2_359', 'P', 'GLU23', 'OE2_291', 'P', 2.9199, 31.5487],
['ASN38', 'N_523', 'P', 'ILE35', 'O_496', 'P', 2.9255, 29.091],
['GLN76', 'NE2_1110', 'P', 'THR46', 'O_657', 'P', 2.9381, 31.3836],
['ARG40', 'NH1_577', 'P', 'THR84', 'OG1_1233', 'P', 2.9482, 8.3748],
['ALA44', 'N_624', 'P', 'PHE10', 'O_103', 'P', 2.9499, 33.1772],
['VAL8', 'N_49', 'P', 'ARG40', 'O_584', 'P', 2.9631, 25.0079],
['ILE35', 'N_478', 'P', 'VAL30', 'O_420', 'P', 2.9811, 23.5092],
['ASN53', 'N_738', 'P', 'GLU50', 'O_711', 'P', 2.995, 28.587],
['ASN34', 'N_464', 'P', 'THR31', 'O_434', 'P', 3.0041, 18.2465],
['ASN15', 'ND2_166', 'P', 'SER43', 'OG_620', 'P', 3.0129, 25.6996],
['ARG27', 'NH1_356', 'P', 'GLU23', 'OE2_291', 'P', 3.0175, 36.9343],
['LEU29', 'N_386', 'P', 'VAL25', 'O_319', 'P', 3.0299, 19.109],
['SER47', 'N_658', 'P', 'LEU13', 'O_149', 'P', 3.0386, 28.8029],
['VAL30', 'N_405', 'P', 'PHE26', 'O_339', 'P', 3.0394, 17.6883],
['ALA24', 'N_294', 'P', 'PRO20', 'O_249', 'P', 3.0751, 29.9487],
['LYS28', 'N_364', 'P', 'ALA24', 'O_303', 'P', 3.0783, 19.9504],
['ALA22', 'N_269', 'P', 'ARG18', 'O_224', 'P', 3.088, 21.873],
['ASP42', 'N_601', 'P', 'VAL8', 'O_64', 'P', 3.1331, 35.5671],
['TRP39', 'N_537', 'P', 'SER36', 'O_507', 'P', 3.1343, 15.1776],
['CYS12', 'N_120', 'P', 'ALA44', 'O_633', 'P', 3.3349, 36.1006]]
Salt Bridges:
In [32]: allRes_20to50[1]
[['ASP32', 'OD1_443_444', 'P', 'LYS28', 'NZ_380', 'P', 3.037],
['ARG27', 'NH1_356_359', 'P', 'GLU23', 'OE1_290_291', 'P', 3.7148],
['ARG75', 'NH1_1090_1093', 'P', 'ASP42', 'OD1_609_610', 'P', 4.5612],
['GLU23', 'OE1_290_291', 'P', 'HSE72', 'NE2_1042', 'P', 4.99]]
We can also select one particular residue or a region of our interest:
In [33]: interactions.getPiCation(selection='resid 85')
[['PHE85',
'1248_1249_1251_1253_1255_1257',
'P',
'ARG40',
'NH1_577_580',
'P',
3.6523]]
In [34]: interactions.getHydrophobic(selection='resid 26 to 100')
[['TYR87', 'OH_1286', 'P', 'ALA156', 'CB_2401', 'P', 3.0459],
['MET63', 'CE_894', 'P', 'ALA24', 'CB_298', 'P', 3.3105],
['ILE68', 'CG2_976', 'P', 'MET63', 'CE_894', 'P', 3.3306],
['LYS6', 'CD_26', 'P', 'TRP39', 'CZ2_555', 'P', 3.5427],
['VAL30', 'CG1_411', 'P', 'PHE26', 'CE2_336', 'P', 3.5603],
['ALA111', 'CB_1677', 'P', 'ILE88', 'CD_1307', 'P', 3.5627],
['VAL11', 'CG2_114', 'P', 'ILE88', 'CG2_1300', 'P', 3.6386],
['VAL41', 'CG2_595', 'P', 'PHE26', 'CD2_334', 'P', 3.6448],
['VAL106', 'CG2_1598', 'P', 'LYS79', 'CG_1155', 'P', 3.6828],
['ILE77', 'CD_1128', 'P', 'LEU99', 'CD2_1480', 'P', 3.6917],
['PHE82', 'CD1_1205', 'P', 'ILE88', 'CD_1307', 'P', 3.692],
['VAL8', 'CG1_55', 'P', 'PHE26', 'CE2_336', 'P', 3.7106],
['MET70', 'CE_1014', 'P', 'MET63', 'CG_890', 'P', 3.7262],
['LEU96', 'CD1_1421', 'P', 'ILE113', 'CG2_1711', 'P', 3.7263],
['LEU9', 'CD2_78', 'P', 'ILE77', 'CD_1128', 'P', 3.745],
['LEU89', 'CD1_1322', 'P', 'VAL8', 'CG2_59', 'P', 3.7672],
['MET91', 'SD_1353', 'P', 'ILE127', 'CD_1949', 'P', 3.8864],
['ALA44', 'CB_628', 'P', 'LEU9', 'CD1_74', 'P', 3.8992],
['ILE21', 'CG2_256', 'P', 'MET63', 'SD_893', 'P', 3.9614],
['LEU153', 'CD1_2350', 'P', 'TRP39', 'NE1_547', 'P', 3.967],
['PHE85', 'CZ_1253', 'P', 'LEU9', 'CD1_74', 'P', 4.0119],
['ILE35', 'CD_491', 'P', 'TRP39', 'NE1_547', 'P', 4.0172],
['LEU29', 'CD1_395', 'P', 'VAL25', 'CG1_310', 'P', 4.0642],
['ALA74', 'CB_1068', 'P', 'ILE16', 'CG2_177', 'P', 4.0772],
['ARG75', 'CG_1081', 'P', 'ALA44', 'CB_628', 'P', 4.0853],
['LYS102', 'CD_1534', 'P', 'ILE77', 'CG2_1121', 'P', 4.1048],
['TYR119', 'CE1_1805', 'P', 'LEU89', 'CD2_1326', 'P', 4.1435],
['ARG40', 'CG_568', 'P', 'PHE85', 'CE2_1257', 'P', 4.2669],
['LYS28', 'CG_371', 'P', 'ILE68', 'CD_983', 'P', 4.2707],
['LYS112', 'CG_1690', 'P', 'TYR87', 'CE1_1283', 'P', 4.3083],
['ARG58', 'CG_820', 'P', 'PHE138', 'CE1_2104', 'P', 4.4781]]
Change selection criteria for interaction type¶
The Interactions.buildInteractionMatrix() method computes interactions
using default parameters for interactions. However, it can be changed
according to our needs. To do that, we need to recalculate the selected type
of interactions using the preferable parameters.
We can do it using the following functions: calcHydrogenBonds(),
calcHydrogenBonds(), calcSaltBridges(),
calcRepulsiveIonicBonding(), calcPiStacking(),
calcPiCation(), calcHydrophohic(),
calcDisulfideBonds(), and use
Interactions.setNewHydrogenBonds(),
Interactions.setNewSaltBridges(),
Interactions.setNewRepulsiveIonicBonding(),
Interactions.setNewPiStacking(),
Interactions.setNewPiCation(),
Interactions.setNewHydrophohic(),
Interactions.setNewDisulfideBonds() method to replace it in the main
Instance.
For example: If we want to replace hydrogen bonds:
In [35]: newHydrogenBonds2 = calcHydrogenBonds(atoms, distA=2.8,
....: angle=30, cutoff_dist=15)
....:
In [36]: interactions.setNewHydrogenBonds(newHydrogenBonds2)
@> Calculating hydrogen bonds.
@> DONOR (res chid atom) <---> ACCEPTOR (res chid atom) Distance Angle
@> GLN143 P NE2_2192 <---> GLU139 P OE2_2126 2.7 9.2
@> HSE66 P NE2_957 <---> GLU139 P OE1_2125 2.7 6.4
@> ARG40 P N_561 <---> LYS6 P O_37 2.7 17.1
@> ARG58 P N_813 <---> ASP56 P OD1_788 2.7 30.0
@> ASN53 P ND2_747 <---> GLU50 P OE1_708 2.8 18.2
@> ALA74 P N_1064 <---> ASN53 P O_751 2.8 21.3
@> ASP56 P N_780 <---> ILE16 P O_189 2.8 27.0
@> Number of detected hydrogen bonds: 7.
@> Hydrogen Bonds are replaced
In [37]: interactions.getHydrogenBonds()
[['GLN143', 'NE2_2192', 'P', 'GLU139', 'OE2_2126', 'P', 2.7287, 9.1823],
['HSE66', 'NE2_957', 'P', 'GLU139', 'OE1_2125', 'P', 2.7314, 6.3592],
['ARG40', 'N_561', 'P', 'LYS6', 'O_37', 'P', 2.7479, 17.1499],
['ARG58', 'N_813', 'P', 'ASP56', 'OD1_788', 'P', 2.7499, 29.9737],
['ASN53', 'ND2_747', 'P', 'GLU50', 'OE1_708', 'P', 2.7702, 18.2336],
['ALA74', 'N_1064', 'P', 'ASN53', 'O_751', 'P', 2.7782, 21.3375],
['ASP56', 'N_780', 'P', 'ILE16', 'O_189', 'P', 2.7793, 27.0481]]
If we want to replace salt bridges, repulsive ionic bonding, or Pi-cation interactions:
In [38]: sb2 = calcSaltBridges(atoms, distA=6)
In [39]: interactions.setNewSaltBridges(sb2)
In [40]: rib2 = calcRepulsiveIonicBonding(atoms, distA=9)
In [41]: interactions.setNewRepulsiveIonicBonding(rib2)
In [42]: picat2 = calcPiCation(atoms, distA=7)
In [43]: interactions.setNewPiCation(picat2)
@> Calculating salt bridges.
@> HSE66 P NE2_957 <---> GLU139 P OE1_2125_2126 2.8
@> ASP81 P OD1_1193_1194 <---> ARG75 P NH1_1090_1093 2.9
@> ASP32 P OD1_443_444 <---> LYS28 P NZ_380 3.0
@> ARG101 P NH1_1516_1519 <---> ASP98 P OD1_1463_1464 3.1
@> ARG27 P NH1_356_359 <---> GLU23 P OE1_290_291 3.7
@> GLU139 P OE1_2125_2126 <---> ARG65 P NH1_938_941 3.8
@> LYS102 P NZ_1540 <---> ASP98 P OD1_1463_1464 3.9
@> ARG58 P NH1_829_832 <---> ASP56 P OD1_788_789 3.9
@> ARG18 P NH1_217_220 <---> ASP92 P OD1_1368_1369 4.1
@> GLU114 P OE1_1735_1736 <---> LYS112 P NZ_1699 4.1
@> ASP120 P OD1_1824_1825 <---> ARG147 P NH1_2257_2260 4.2
@> LYS110 P NZ_1667 <---> ASP86 P OD1_1269_1270 4.2
@> GLU114 P OE1_1735_1736 <---> HSE157 P NE2_2418 4.4
@> ARG18 P NH1_217_220 <---> ASP129 P OD1_1978_1979 4.6
@> ARG75 P NH1_1090_1093 <---> ASP42 P OD1_609_610 4.6
@> GLU23 P OE1_290_291 <---> HSE72 P NE2_1042 5.0
@> ASP42 P OD1_609_610 <---> HSE72 P NE2_1042 5.4
@> ASP81 P OD1_1193_1194 <---> ARG40 P NH1_577_580 5.8
@> Number of detected salt bridges: 18.
@> Salt Bridges are replaced
@> Calculating repulsive ionic bonding.
@> ASP42 P OD1_609_610 <---> ASP81 P OD1_1193_1194 6.7
@> GLU80 P OE1_1181_1182 <---> ASP81 P OD1_1193_1194 7.0
@> ASP92 P OD1_1368_1369 <---> ASP129 P OD1_1978_1979 7.6
@> LYS110 P NZ_1667 <---> ARG40 P NH1_577_580 7.8
@> ASP92 P OD1_1368_1369 <---> GLU93 P OE1_1383_1384 8.6
@> GLU128 P OE1_1966_1967 <---> ASP137 P OD1_2090_2091 8.9
@> Number of detected Repulsive Ionic Bonding interactions: 6.
@> Repulsive Ionic Bonding are replaced
@> Calculating cation-Pi interactions.
@> PHE85 P 1248_1249_1251_1253_1255_1257 <---> ARG40 P NH1_577_580 3.7
@> HSE66 P 953_954_955_957_959 <---> ARG65 P NH1_938_941 4.5
@> HSE157 P2414_2415_2416_2418_2420_2423_2424 <---> LYS112 P NZ_1699 4.8
@> PHE138 P 2101_2102_2104_2106_2108_2110 <---> ARG58 P NH1_829_832 5.0
@> TYR131 P 2003_2004_2006_2008_2011_2013 <---> ARG58 P NH1_829_832 5.1
@> PHE85 P 1248_1249_1251_1253_1255_1257 <---> ARG75 P NH1_1090_1093 6.3
@> TRP39 P 549_550_551_553_555_557 <---> LYS6 P NZ_32 6.6
@> TYR87 P 1280_1281_1283_1285_1288_1290 <---> LYS112 P NZ_1699 6.8
@> Number of detected cation-pi interactions: 8.
@> Pi-Cation interactions are replaced
Assess the functional significance of a residue¶
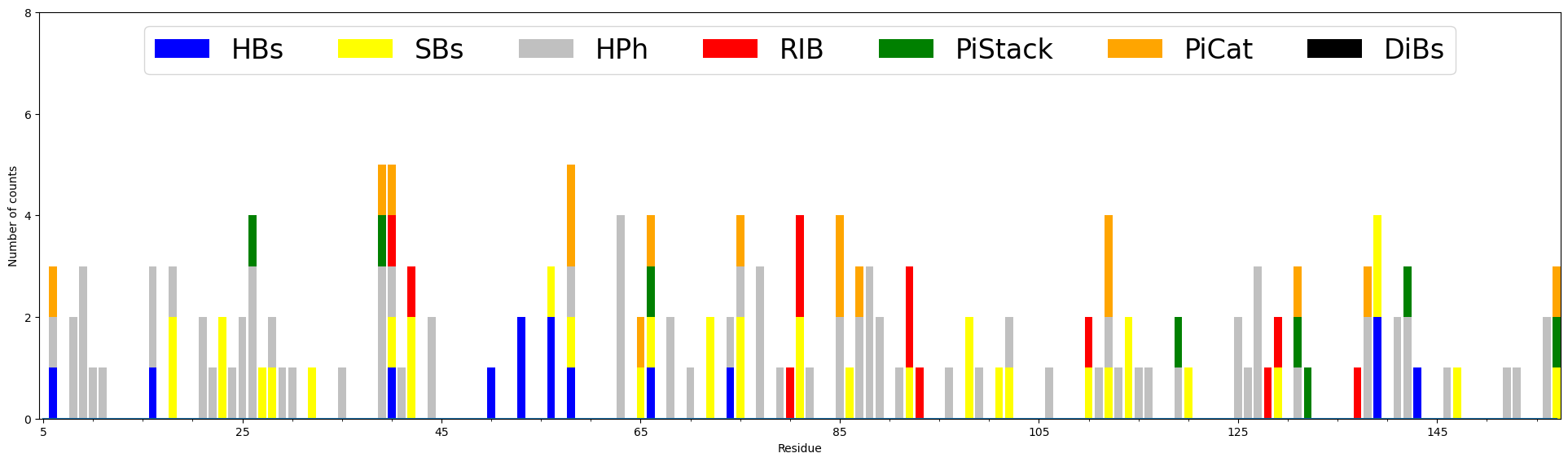
For assessing the functional significance of each residue in protein structure, we counted the number of possible contacts based on:
- Hydrogen bonds (
HBs)- Salt Bridges (
SBs)- Repulsive Ionic Bonding (
RIB)- Pi stacking interactions (
PiStack)- Pi-cation interactions (
PiCat)- Hydrophobic interactions (
HPh)- Disulfide Bonds (
DiBs)
To compute the weighted interactions use the
Interactions.buildInteractionMatrix() method:
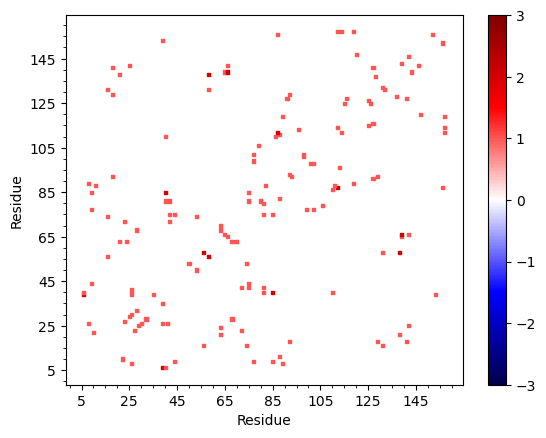
In [44]: matrix = interactions.buildInteractionMatrix()
@> Calculating interactions
The results can be displayed in the following way:
In [45]: import matplotlib.pylab as plt
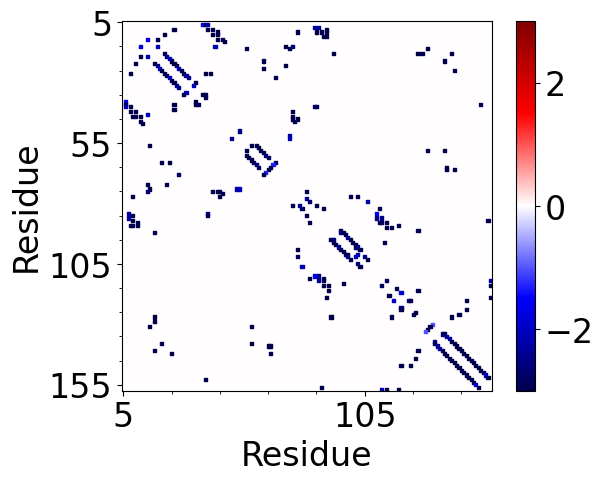
In [46]: showAtomicMatrix(matrix, atoms=atoms.ca, cmap='seismic', markersize=5)
In [47]: plt.xlabel('Residue')
In [48]: plt.ylabel('Residue')
In [49]: plt.clim([-3,3])
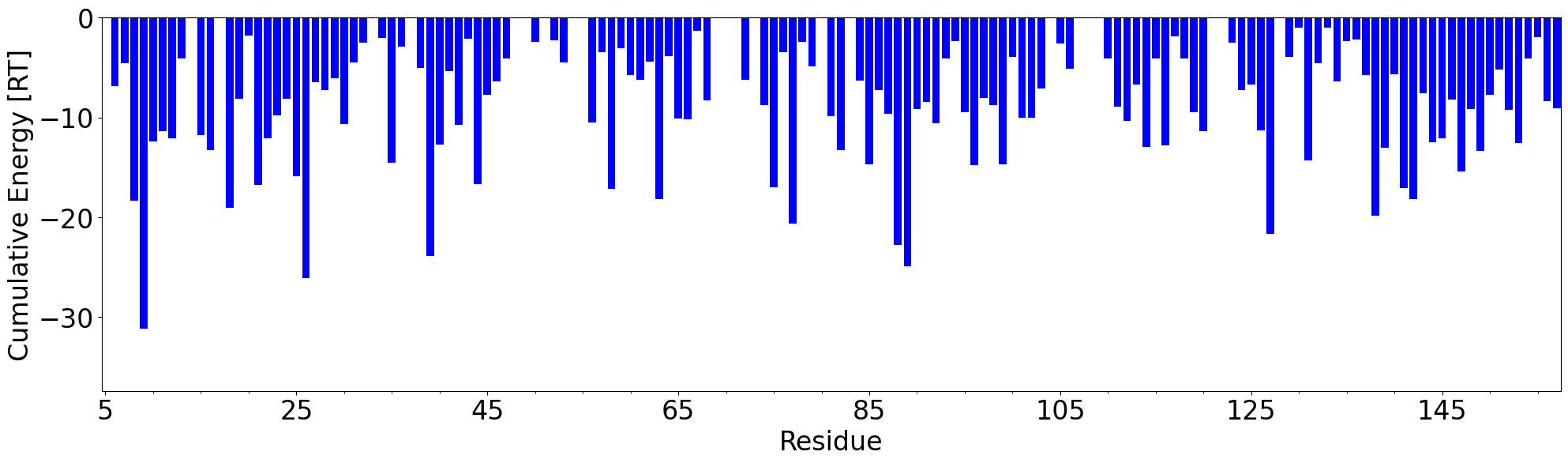
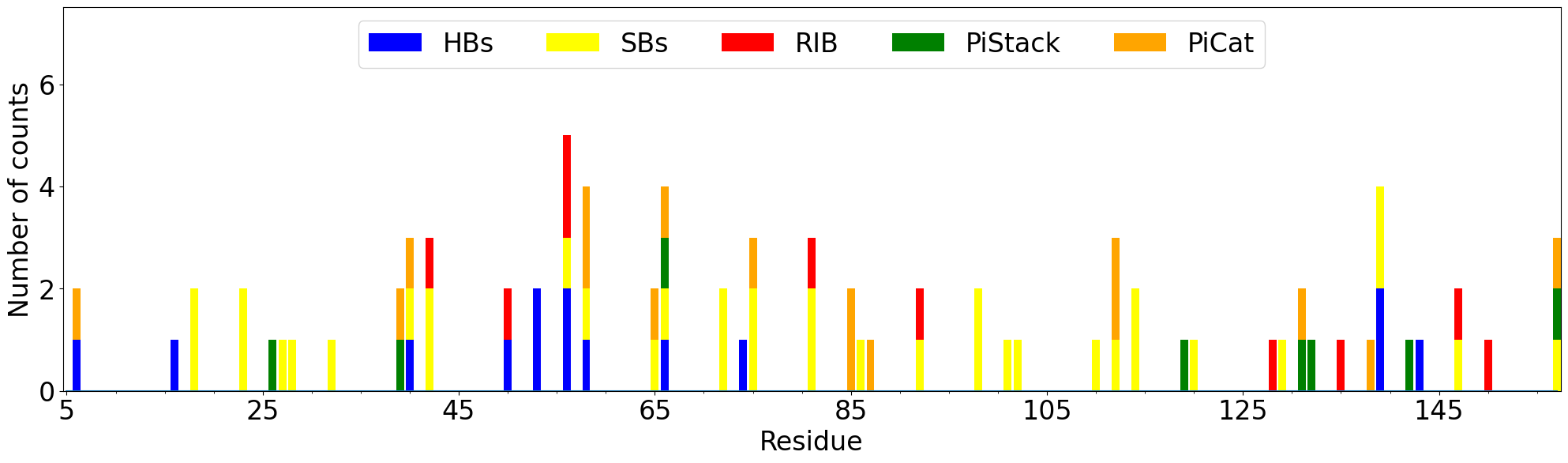
The total number of interaction for each residue can be displayed on the plot using
showCumulativeInteractionTypes() function.
In [50]: interactions.showCumulativeInteractionTypes()
@> Calculating interaction matrix
@> Calculating interaction matrix
@> Calculating interaction matrix
@> Calculating interaction matrix
@> Calculating interaction matrix
@> Calculating interaction matrix
@> Calculating interaction matrix
Energy of interactions¶
Additionally, we can also obtain energy of the pairs of identified interactions
with several functions. One of them is showPairEnergy() method that will
check the energy of interactions for pairs of residues and add it to the
interactions list. Energies will be added at the last position for each
pair.
In [51]: showPairEnergy(interactions.getHydrogenBonds())
[['ARG101', 'NH1_1516', 'P', 'ASP98', 'OD1_1463', 'P', 1.998, 33.1238, -3.92],
['HIS72', 'NE2_1042', 'P', 'ASN15', 'OD1_165', 'P', 2.5997, 34.752, -3.05],
['GLN143', 'NE2_2192', 'P', 'GLU139', 'OE2_2126', 'P', 2.7287, 9.1823, -3.45],
['HIS66', 'NE2_957', 'P', 'GLU139', 'OE1_2125', 'P', 2.7314, 6.3592, -3.15],
['ARG40', 'N_561', 'P', 'LYS6', 'O_37', 'P', 2.7479, 17.1499, -2.11],
['ARG58', 'N_813', 'P', 'ASP56', 'OD1_788', 'P', 2.7499, 29.9737, -3.92],
['ALA45', 'N_634', 'P', 'ARG75', 'O_1097', 'P', 2.7609, 35.0983, -3.26],
['ASN53', 'ND2_747', 'P', 'GLU50', 'OE1_708', 'P', 2.7702, 18.2336, -2.43],
['ALA74', 'N_1064', 'P', 'ASN53', 'O_751', 'P', 2.7782, 21.3375, -2.06],
['ASP56', 'N_780', 'P', 'ILE16', 'O_189', 'P', 2.7793, 27.0481, -3.46],
['LYS110', 'NZ_1667', 'P', 'THR84', 'O_1240', 'P', 2.7977, 38.2213, -1.64],
['LEU116', 'N_1758', 'P', 'CYS90', 'O_1342', 'P', 2.8072, 15.0239, -6.13],
['SER103', 'N_1546', 'P', 'LEU99', 'O_1485', 'P', 2.8075, 29.107, -4.12],
['ASN134', 'N_2045', 'P', 'ASP137', 'OD2_2091', 'P', 2.8132, 22.562, -2.5],
['PHE152', 'N_2321', 'P', 'CYS148', 'O_2275', 'P', 2.8141, 8.2562, -4.81],
['ASN95', 'N_1398', 'P', 'ASP92', 'OD1_1368', 'P', 2.8148, 12.5701, -2.5],
['LYS6', 'N_16', 'P', 'ASN38', 'O_536', 'P', 2.8178, 25.0305, -1.63],
['ILE77', 'N_1115', 'P', 'ALA45', 'O_643', 'P', 2.8179, 12.1855, -4.45],
['ARG58', 'NH2_832', 'P', 'ASP56', 'OD2_789', 'P', 2.8204, 27.6617, -3.92],
['LEU99', 'N_1467', 'P', 'ASN95', 'O_1411', 'P', 2.8205, 15.4867, -3.94],
['CYS149', 'N_2276', 'P', 'CYS145', 'O_2224', 'P', 2.8247, 9.5914, -7.23],
['GLY52', 'N_731', 'P', 'ALA74', 'O_1073', 'P', 2.832, 6.6442, -2.24],
['ASP32', 'N_435', 'P', 'LYS28', 'O_385', 'P', 2.8357, 8.8318, -2.47],
['ILE88', 'N_1294', 'P', 'LYS112', 'O_1704', 'P', 2.8429, 17.7147, -2.88],
..
..
Another function is Interactions.buildInteractionMatrixEnergy() which
provides matrix with energies of interactions for each pair.
In [52]: matrix_en = interactions.buildInteractionMatrixEnergy()
@> Calculating interaction matrix
The results can be displayed in the following way:
In [53]: import matplotlib.pylab as plt
In [54]: showAtomicMatrix(matrix_en, atoms=atoms.ca, cmap='seismic', markersize=5)
In [55]: plt.xlabel('Residue')
In [56]: plt.ylabel('Residue')
In [57]: plt.clim([-3,3])
The total energy of interaction for each residue can be displayed on the plot using
showCumulativeInteractionTypes() function with energy=True.
In [58]: interactions.showCumulativeInteractionTypes(energy=True)
@> Calculating interaction matrix
@> Calculating interaction matrix
@> Calculating interaction matrix
@> Calculating interaction matrix
@> Calculating interaction matrix
@> Calculating interaction matrix
@> Calculating interaction matrix
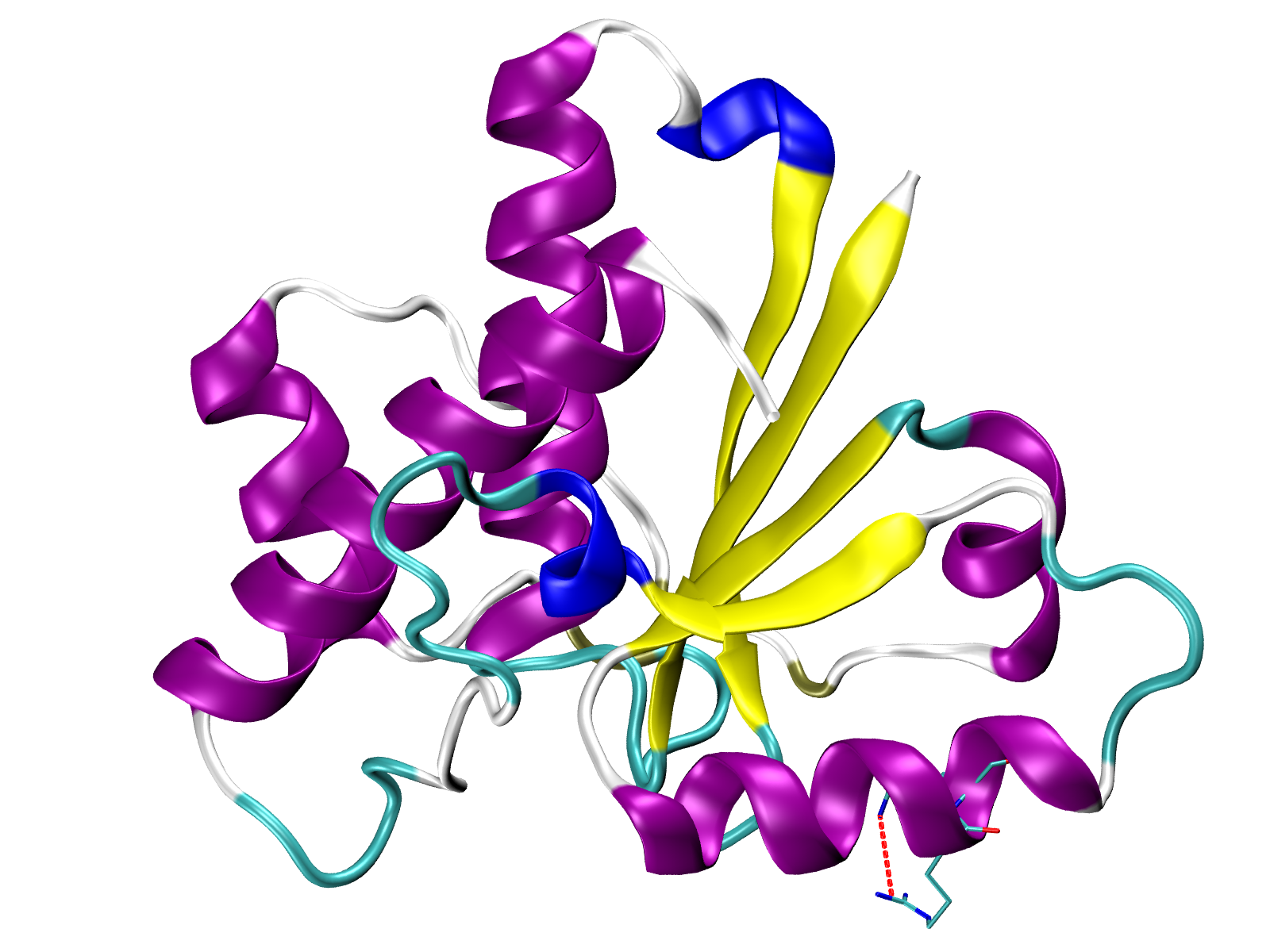
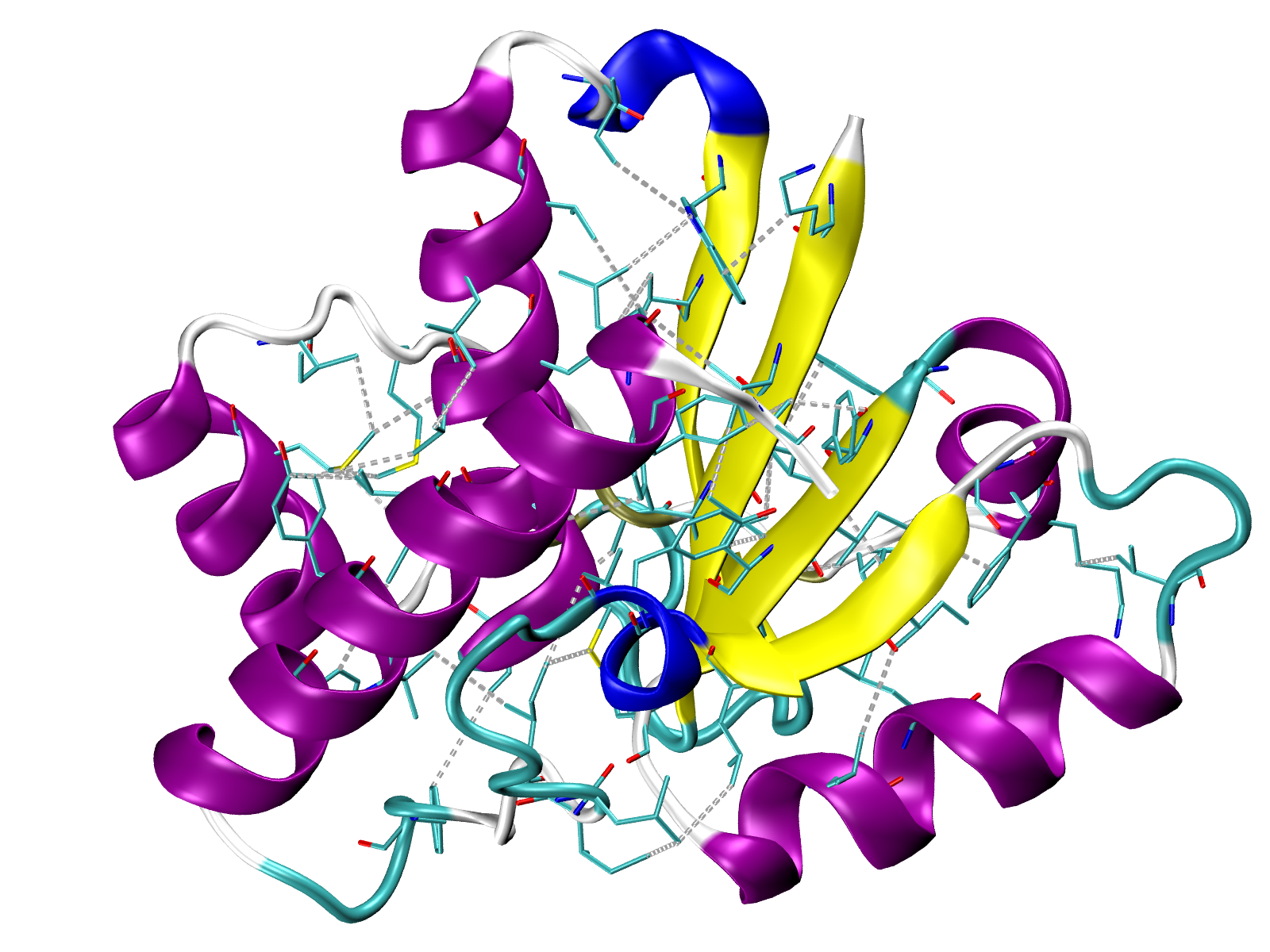
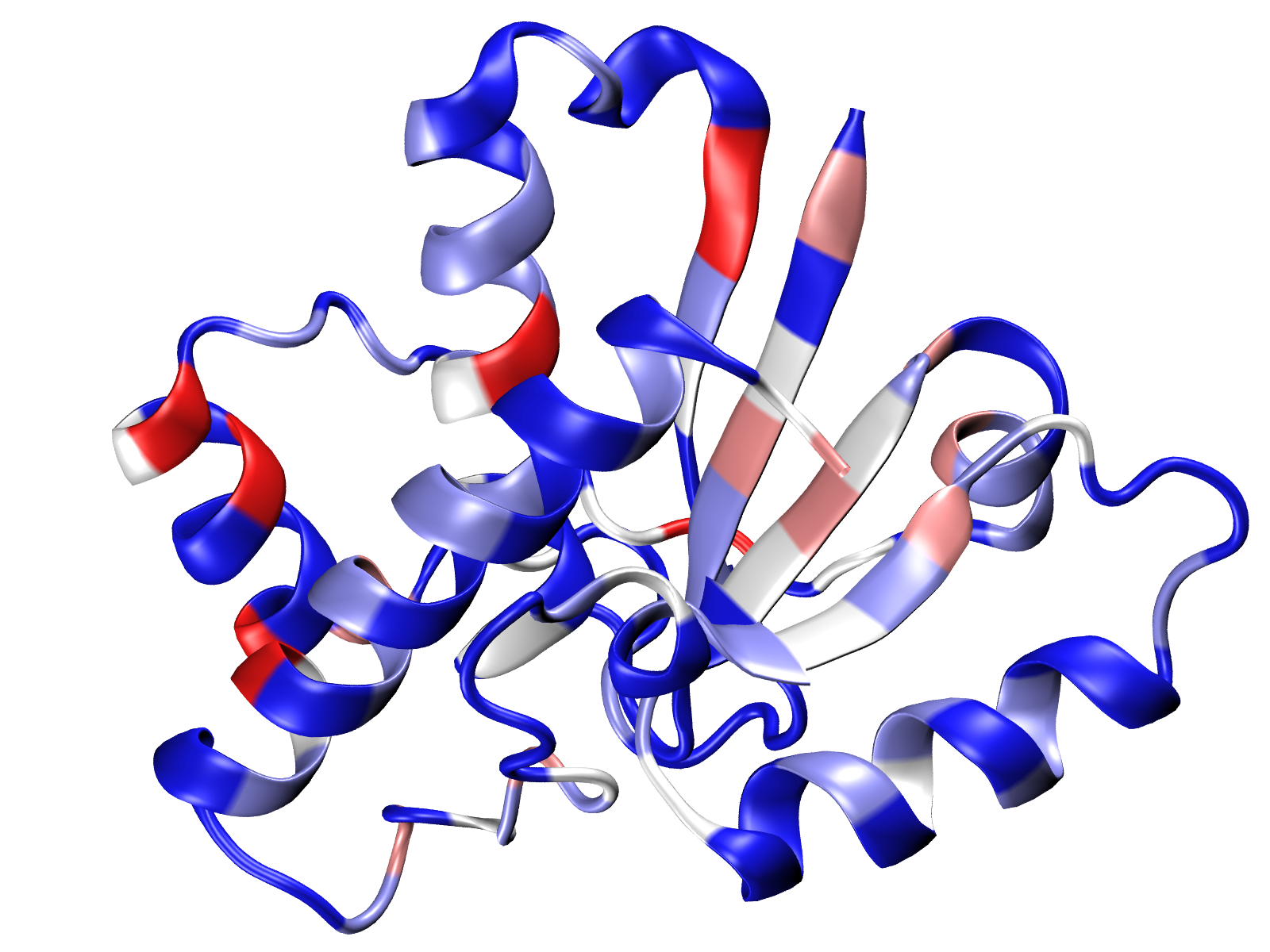
Visualize number of interactions onto 3D structure¶
The number of the interaction or total energy can be saved to a PDB file in the
Occupancy column by using Interactions.saveInteractionsPDB()
method. Then the score would be displayed in color in any available graphical
program, for example, in VMD.
In [59]: interactions.saveInteractionsPDB(filename='5kqm_meanMatrix.pdb')
@> PDB file saved.
A file 5kqm_meanMatrix.pdb will be saved and can be used in VMD by
uploading PDB structure and displaying it with Coloring Method
Occupancy. By default blue colors correspond to the highest
values but we can change it in .
To save energy of interaction instead of number of interactions we can use the following command:
In [60]: interactions.saveInteractionsPDB(filename='5kqm_meanMatrix_en.pdb', energy=True)
@> PDB file saved.
Exclude some interaction types from calculations¶
For analysis we can exclude some of the interaction types by assigning zero
to the type of interactions (HBs - hydrogen bonds, SBs - salt bridges, RIB -
repulsive ionic bonding, PiCat - Pi-Cation, PiStack - Pi-Stacking, HPh -
hydrophobic interactions and finally DiBs - disulfide bonds).
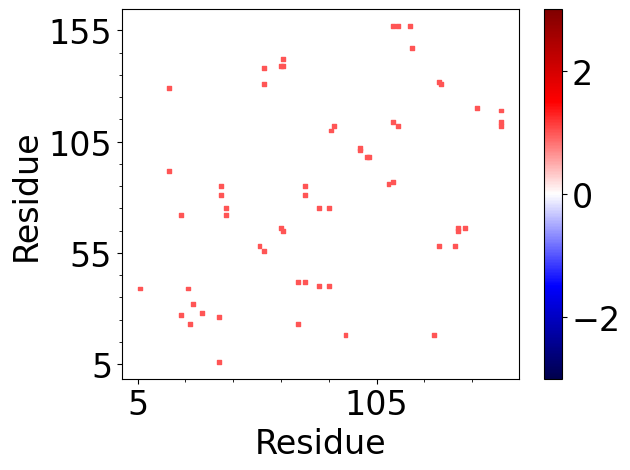
In [61]: matrix = interactions.buildInteractionMatrix(RIB=0, HBs=0, HPh=0, DiBs=0)
@> Calculating interaction matrix
The results can be displayed in a similar way:
In [62]: showAtomicMatrix(matrix, atoms=atoms.ca, cmap='seismic', markersize=8)
In [63]: plt.xlabel('Residue')
In [64]: plt.ylabel('Residue')
In [65]: plt.clim([-3,3])
In [66]: interactions.showCumulativeInteractionTypes(HPh=0, DiBs=0)
@> Calculating interaction matrix
@> Calculating interaction matrix
@> Calculating interaction matrix
@> Calculating interaction matrix
@> Calculating interaction matrix
@> Calculating interaction matrix
@> Calculating interaction matrix