PDB structure with multiple chains¶
This time we will use protein with two chains, lipoxygenase (PDB: 7LAF) which contain chain A and chain B. First, we will add missing hydrogens to the protein structures and then we will perform analysis of interactions between two chains.
Add missing hydrogen atoms to the structure¶
We start by fetching the PDB file and adding missing hydrogens using Openbabel.
In [1]: fetchPDB('7laf', compressed=False)
In [2]: addMissingAtoms('7laf.pdb', method='openbabel')
In [3]: atoms = parsePDB('addH_7laf.pdb').select('protein')
@> Connecting wwPDB FTP server RCSB PDB (USA).
@> 7laf downloaded (7laf.pdb)
@> PDB download via FTP completed (1 downloaded, 0 failed).
@> Hydrogens were added to the structure. Structure addH_7laf.pdb is saved in the local directry.
@> 21970 atoms and 1 coordinate set(s) were parsed in 0.24s.
Perform InSty calculations and extract chain-chain interactions¶
In [4]: interactions = Interactions('7laf')
Compute Interactions¶
To compute all interactions:
In [5]: all_interactions = interactions.calcProteinInteractions(atoms)
@> Calculating interations.
@> Calculating hydrogen bonds.
@> DONOR (res chid atom) <---> ACCEPTOR (res chid atom) Distance Angle
@> ASP505 A OD2_3935 <---> TYR496 A OH_3867 2.2 29.5
@> ASP666 B OD1_10390 <---> SER648 B OG_10241 2.3 17.8
@> HIS394 A N_3026 <---> GLU141 A OE2_1006 2.4 35.5
@> ARG390 B NH1_8204 <---> TYR149 B O_6273 2.4 35.6
@> GLN641 A NE2_4984 <---> GLY621 A O_4815 2.4 8.5
@> ARG649 B NH1_10251 <---> GLU653 B OE1_10281 2.4 4.9
@> ARG463 B NH1_8796 <---> ASP459 B OD1_8759 2.4 30.6
@> TYR318 B N_7621 <---> LEU327 B O_7687 2.4 4.3
@> ASN301 A ND2_2299 <---> ASP428 A O_3307 2.4 29.3
@> ARG474 A NH1_3686 <---> ILE468 A O_3625 2.5 37.5
@> ARG474 B NH1_8890 <---> VAL465 B O_8805 2.5 21.1
@> SER517 B OG_9247 <---> ASN522 B ND2_9287 2.5 26.7
@> ASN522 B ND2_9287 <---> SER517 B OG_9247 2.5 33.0
@> TRP481 B N_8937 <---> GLY477 B O_8911 2.5 7.9
@> LEU36 A N_274 <---> VAL24 A O_194 2.5 21.5
@> SER526 A OG_4113 <---> GLN523 A O_4087 2.5 24.9
@> ARG138 B NH2_6183 <---> GLU507 B OE1_9158 2.5 28.0
@> TRP109 A N_743 <---> ASN173 A OD1_1279 2.5 7.1
@> THR431 B OG1_8538 <---> VAL427 B O_8504 2.5 24.5
@> SER501 B OG_9107 <---> SER498 B O_9082 2.5 35.7
@> GLN143 B N_6219 <---> GLN139 B O_6187 2.5 11.9
@> LEU329 A N_2495 <---> LEU316 A O_2404 2.5 21.1
@> LEU110 A N_757 <---> TRP87 A O_551 2.6 33.6
@> HIS373 A N_2854 <---> HIS368 A O_2818 2.6 19.3
@> ASN413 B ND2_8402 <---> GLU382 B OE1_8140 2.6 20.3
@> ARG535 B NH1_9382 <---> TYR496 B O_9063 2.6 29.4
@> ARG649 A NH1_5047 <---> GLU653 A OE2_5078 2.6 19.6
@> TRP109 B N_5947 <---> ASN173 B OD1_6483 2.6 37.3
@> ARG429 B N_8516 <---> GLN425 B O_8488 2.6 23.9
@> GLN575 B NE2_9683 <---> LEU594 B O_9812 2.6 14.9
@> ARG68 A N_482 <---> SER25 A O_201 2.6 14.5
@> HIS70 A N_500 <---> SER23 A O_188 2.6 12.3
@> ARG215 A NH2_1620 <---> GLU168 B OE1_6442 2.6 24.8
@> THR462 A N_3576 <---> GLU458 A O_3543 2.6 22.5
@> ARG463 B NH2_8797 <---> TYR471 B OH_8856 2.6 19.7
@> HIS405 A ND1_3124 <---> ASN672 A O_5230 2.6 25.8
@> GLN335 A NE2_2547 <---> ALA312 A O_2377 2.6 10.5
@> LEU110 B N_5961 <---> TRP87 B O_5770 2.6 37.7
@> TYR495 B OH_9059 <---> GLU630 B OE1_10098 2.6 12.0
@> ARG463 A NH2_3593 <---> ILE251 A O_1918 2.6 39.3
@> GLN319 B NE2_7641 <---> GLY324 B O_7668 2.6 28.2
@> HIS373 B N_8058 <---> HIS368 B O_8022 2.6 20.9
@> GLU364 B N_7982 <---> VAL360 B O_7954 2.6 28.1
@> TRP87 A N_548 <---> LEU110 A O_760 2.6 8.9
@> TYR408 A OH_3157 <---> ASP616 A OD1_4775 2.6 37.7
@> GLY493 B N_9036 <---> SER489 B O_9008 2.6 32.4
@> LEU354 B N_7900 <---> LYS350 B O_7858 2.6 22.3
@> LEU142 A N_1007 <---> ARG138 A O_972 2.6 5.8
@> VAL488 A N_3794 <---> VAL484 A O_3759 2.6 11.4
@> ASN655 A ND2_5097 <---> TYR662 A O_5144 2.6 17.4
@> THR95 A N_632 <---> ARG5 A O_52 2.6 12.4
@> ARG208 A N_1551 <---> GLU212 A OE1_1591 2.6 21.7
@> ARG463 A NH2_3593 <---> TYR471 A OH_3652 2.6 35.2
@> ARG208 B N_6755 <---> GLU212 B OE1_6795 2.6 34.4
@> SER550 B OG_9500 <---> ILE546 B O_9466 2.6 22.5
@> GLN119 B NE2_6029 <---> GLN137 B OE1_6171 2.6 28.1
@> LEU327 B N_7684 <---> TYR318 B O_7624 2.6 6.3
@> LEU420 A N_3247 <---> ALA416 A O_3217 2.6 34.5
@> CYS106 A N_716 <---> ARG90 A O_582 2.7 36.5
@> LEU607 B N_9900 <---> VAL603 B O_9875 2.7 12.6
@> VAL488 B N_8998 <---> VAL484 B O_8963 2.7 4.8
@> GLY583 B N_9735 <---> ASP352 B OD2_7885 2.7 15.8
@> SER25 A N_198 <---> ARG68 A O_485 2.7 38.0
@> ASN301 A ND2_2299 <---> THR431 A O_3332 2.7 28.3
@> ARG407 A NH2_3145 <---> ASP616 A O_4772 2.7 29.7
@> GLN509 B NE2_9176 <---> LEU532 B O_9352 2.7 37.1
@> ARG407 A NH1_3144 <---> ASP616 A OD2_4776 2.7 11.0
@> SER489 A N_3801 <---> GLU485 A O_3766 2.7 24.6
@> ARG215 A NH1_1619 <---> GLU168 B OE2_6443 2.7 28.7
@> ARG253 A N_1934 <---> ARG463 A O_3586 2.7 10.7
@> PHE288 B N_7398 <---> LEU317 B O_7616 2.7 18.1
@> GLN509 A NE2_3972 <---> LEU532 A O_4148 2.7 27.8
@> THR409 A OG1_3163 <---> VAL674 A O_5244 2.7 37.9
@> PHE309 B N_7556 <---> MET574 B O_9670 2.7 12.8
@> ASN445 B N_8639 <---> LEU441 B O_8606 2.7 23.6
@> GLY583 A N_4531 <---> ASP352 A OD2_2681 2.7 14.6
@> TYR451 B N_8689 <---> SER526 B O_9315 2.7 38.7
@> ARG5 A N_49 <---> THR95 A O_635 2.7 15.9
@> CYS106 B N_5920 <---> ARG90 B O_5801 2.7 32.5
@> TYR149 A N_1066 <---> ARG145 A O_1032 2.7 8.1
@> GLN575 A N_4471 <---> THR593 A O_4601 2.7 15.0
@> HIS160 A N_1167 <---> LYS518 A O_4047 2.7 35.5
@> PHE547 B N_9471 <---> THR543 B O_9444 2.7 33.0
@> ARG253 B N_7138 <---> ARG463 B O_8790 2.7 16.2
@> ASN655 A N_5090 <---> ILE651 A O_5056 2.7 28.0
@> LEU345 B N_7817 <---> ASP348 B OD2_7846 2.7 14.0
@> ASP504 A N_3920 <---> GLU500 A O_3892 2.7 37.2
@> ARG203 A NH1_1514 <---> GLU212 A OE2_1592 2.7 38.1
@> ASN569 A ND2_4434 <---> SER563 A O_4384 2.7 2.5
@> TRP481 A N_3733 <---> GLY477 A O_3707 2.7 5.8
@> ASN362 A N_2765 <---> THR358 A O_2729 2.7 34.3
@> MET314 A N_2386 <---> GLN332 A O_2519 2.7 23.3
@> SER430 B N_8527 <---> VAL426 B O_8497 2.7 13.0
@> TRP87 B N_5767 <---> LEU110 B O_5964 2.7 31.2
@> HIS368 B N_8019 <---> GLU364 B O_7985 2.7 24.6
@> ILE492 B N_9028 <---> VAL488 B O_9001 2.7 2.1
@> ASN413 A ND2_3198 <---> HIS378 A O_2899 2.7 35.7
@> ARG390 A NH1_3000 <---> TYR149 A O_1069 2.7 18.4
@> ARG407 B NH2_8349 <---> ASP616 B O_9976 2.7 31.8
@> SER430 A N_3323 <---> VAL426 A O_3293 2.7 37.6
@> ARG654 A N_5079 <---> GLY650 A O_5052 2.7 22.2
@> ASN445 A N_3435 <---> LEU441 A O_3402 2.7 9.8
@> HIS376 B N_8084 <---> LEU371 B O_8046 2.7 15.5
@> LYS518 B N_9248 <---> GLU514 B O_9217 2.7 30.5
@> ARG444 B NH2_8638 <---> SER296 B O_7465 2.7 14.5
@> GLU440 A N_3390 <---> GLU436 A O_3363 2.7 23.1
@> LEU607 A N_4696 <---> VAL603 A O_4671 2.7 6.3
@> GLN641 A N_4976 <---> ILE637 A O_4948 2.7 35.5
@> ARG444 B N_8628 <---> GLU440 B O_8597 2.7 20.9
@> ASP202 A OD2_1504 <---> GLU418 B OE2_8442 2.7 31.6
@> ILE403 B N_8307 <---> PHE399 B O_8274 2.7 13.4
@> LEU278 A N_2119 <---> ASP265 A OD1_2043 2.7 32.7
@> GLN575 B N_9675 <---> THR593 B O_9805 2.7 22.3
@> GLN136 A NE2_959 <---> GLU140 A OE2_997 2.7 16.6
@> TYR496 B N_9060 <---> ILE492 B O_9031 2.7 35.5
@> GLU364 A N_2778 <---> VAL360 A O_2750 2.8 29.9
@> ALA188 A N_1398 <---> PHE184 A O_1361 2.8 22.9
@> ASN672 B N_10431 <---> ARG618 B O_9993 3.3 14.1
@> ARG461 A N_3565 <---> PRO457 A O_3536 3.3 31.9
@> SER636 A N_4939 <---> ALA632 A O_4908 3.3 26.8
@> GLN136 B NE2_6163 <---> GLU140 B OE2_6201 3.3 15.4
@> ALA370 A N_2834 <---> SER366 A O_2801 3.3 16.7
@> VAL360 A N_2747 <---> ALA356 A O_2715 3.3 23.8
@> PHE229 A N_1729 <---> ALA225 A O_1703 3.3 33.3
@> ASN362 A ND2_2772 <---> PRO571 A O_4446 3.3 10.2
@> CYS161 A N_1177 <---> LYS152 A O_1104 3.3 8.3
@> ALA370 B N_8038 <---> SER366 B O_8005 3.3 30.9
@> ASN413 B N_8395 <---> THR409 B O_8365 3.3 36.3
@> THR372 A N_2847 <---> PHE367 A O_2807 3.3 18.6
@> ARG215 B NH1_6823 <---> GLU212 B OE2_6796 3.3 34.9
@> ASN598 B ND2_9845 <---> ASN304 B OD1_7525 3.3 7.3
@> GLY424 B N_8481 <---> ASP428 B OD2_8515 3.3 25.2
@> ILE515 B N_9223 <---> TRP511 B O_9185 3.3 19.4
@> ARG361 A NH1_2763 <---> ASN569 A O_4430 3.3 27.9
@> CYS161 B N_6381 <---> LYS152 B O_6308 3.3 19.0
@> THR95 B OG1_5856 <---> ARG5 B O_5309 3.3 14.8
@> SER517 B OG_9247 <---> ASN522 B OD1_9286 3.3 36.6
@> ARG474 B NH1_8890 <---> ILE468 B N_8826 3.3 28.8
@> VAL268 A N_2058 <---> THR264 A O_2033 3.3 27.6
@> SER377 B N_8094 <---> THR372 B O_8054 3.3 31.9
@> ARG535 A N_4169 <---> ASP499 A OD1_3887 3.3 1.1
@> ARG634 B NH2_10131 <---> GLU626 B OE1_10061 3.3 31.2
@> ILE421 B N_8459 <---> ALA416 B O_8421 3.3 24.8
@> THR10 A N_91 <---> ALA49 A O_335 3.3 21.1
@> TYR473 B N_8869 <---> ASN244 B OD1_7075 3.3 39.9
@> GLN241 A NE2_1845 <---> ASN569 A OD1_4433 3.3 38.6
@> TYR495 A N_3844 <---> ILE491 A O_3819 3.3 30.8
@> ILE421 A N_3255 <---> ALA416 A O_3217 3.3 34.5
@> ARG5 B N_5306 <---> THR95 B O_5854 3.3 34.8
@> GLN139 A N_980 <---> GLN135 A O_945 3.4 11.3
@> GLN479 A N_3716 <---> ASP475 A O_3691 3.4 33.6
@> SER286 B N_7384 <---> GLU281 B O_7348 3.4 16.8
@> MET195 B N_6647 <---> ALA191 B O_6620 3.4 27.9
@> ARG618 A NH1_4795 <---> ASP625 A OD2_4849 3.4 9.3
@> VAL502 B N_9108 <---> SER498 B O_9082 3.4 32.3
@> ILE515 A N_4019 <---> TRP511 A O_3981 3.4 28.7
@> ARG407 B NH2_8349 <---> GLU671 B OE2_10430 3.4 26.5
@> ASN672 A N_5227 <---> ARG618 A O_4789 3.4 31.4
@> VAL167 B N_6428 <---> GLU418 B OE1_8441 3.4 23.0
@> SER320 B N_7642 <---> PRO325 B O_7672 3.4 32.0
@> HIS394 B N_8230 <---> GLU141 B OE1_6209 3.4 4.5
@> ARG203 A NH1_1514 <---> GLU212 A OE1_1591 3.4 24.9
..
..
@> Number of detected hydrogen bonds: 669.
@> Calculating salt bridges.
@> LYS196 A NZ_1459 <---> ASP202 A OD1_1503_1504 2.4
@> GLU168 B OE1_6442_6443 <---> ARG215 A NH1_1619_1620 2.6
@> ASP202 B OD1_6707_6708 <---> LYS196 B NZ_6663 2.7
@> ARG654 A NH1_5088_5089 <---> ASP476 A OD1_3702_3703 2.8
@> ASP505 B OD1_9138_9139 <---> HIS396 B NE2_8255 2.9
@> ARG203 A NH1_1514_1515 <---> GLU212 A OE1_1591_1592 3.0
@> GLU281 B OE1_7352_7353 <---> LYS284 B NZ_7379 3.0
@> ASP616 A OD1_4775_4776 <---> ARG407 A NH1_3144_3145 3.0
@> ASP505 A OD1_3934_3935 <---> HIS396 A NE2_3051 3.0
@> LYS582 B NZ_9734 <---> ASP348 B OD1_7845_7846 3.1
@> ARG635 A NH1_4937_4938 <---> GLU631 A OE1_4903_4904 3.2
@> GLU32 B OE1_5509_5510 <---> ARG68 B NH1_5729_5730 3.3
@> GLU212 B OE1_6795_6796 <---> ARG203 B NH1_6718_6719 3.3
@> ASP625 B OD1_10052_10053 <---> ARG618 B NH1_9999_10000 3.3
@> ASP616 B OD1_9979_9980 <---> ARG407 B NH1_8348_8349 3.3
@> HIS292 A NE2_2237 <---> GLU364 A OE1_2785_2786 3.4
@> ARG618 A NH1_4795_4796 <---> ASP625 A OD1_4848_4849 3.4
@> ASP476 B OD1_8906_8907 <---> ARG654 B NH1_10292_10293 3.5
@> ARG138 B NH1_6182_6183 <---> GLU507 B OE1_9158_9159 3.5
@> ARG649 B NH1_10251_10252 <---> GLU653 B OE1_10281_10282 3.6
@> ARG649 A NH1_5047_5048 <---> GLU653 A OE1_5077_5078 3.6
@> ARG634 B NH1_10130_10131 <---> GLU626 B OE1_10061_10062 3.7
@> GLU364 B OE1_7989_7990 <---> HIS292 B NE2_7441 3.7
@> ARG220 B NH1_6872_6873 <---> GLU194 B OE1_6645_6646 3.8
@> GLU507 A OE1_3954_3955 <---> ARG138 A NH1_978_979 3.8
@> ASP602 A OD1_4666_4667 <---> ARG429 A NH1_3321_3322 3.9
@> GLU626 A OE1_4857_4858 <---> ARG634 A NH1_4926_4927 3.9
@> ARG220 A NH1_1668_1669 <---> GLU194 A OE1_1441_1442 3.9
@> LYS357 B NZ_7929 <---> ASP235 B OD1_7001_7002 3.9
@> LYS175 A NZ_1297 <---> GLU168 A OE1_1238_1239 4.0
@> ASP235 A OD1_1797_1798 <---> LYS357 A NZ_2725 4.0
@> GLU141 B OE1_6209_6210 <---> ARG145 B NH1_6242_6243 4.0
@> ARG429 B NH1_8525_8526 <---> ASP602 B OD1_9870_9871 4.0
@> GLU613 A OE1_4756_4757 <---> LYS180 A NZ_1336 4.0
@> ARG7 A NH1_76_77 <---> ASP52 A OD1_361_362 4.1
@> ARG463 B NH1_8796_8797 <---> ASP459 B OD1_8759_8760 4.1
@> GLU382 A OE1_2936_2937 <---> ARG417 A NH1_3228_3229 4.1
@> ASP348 A OD1_2641_2642 <---> LYS582 A NZ_4530 4.2
@> ASP20 B OD1_5424_5425 <---> LYS71 B NZ_5756 4.2
@> GLU194 A OE1_1441_1442 <---> LYS198 A NZ_1476 4.2
@> GLU32 A OE1_252_253 <---> ARG68 A NH1_491_492 4.3
@> ARG463 A NH1_3592_3593 <---> ASP459 A OD1_3555_3556 4.3
@> ARG208 A NH1_1560_1561 <---> GLU111 B OE1_5976_5977 4.3
@> GLU141 A OE1_1005_1006 <---> ARG145 A NH1_1038_1039 4.4
@> ASP475 A OD1_3694_3695 <---> ARG474 A NH1_3686_3687 4.4
@> ASP616 A OD1_4775_4776 <---> LYS180 A NZ_1336 4.5
@> ARG390 A NH1_3000_3001 <---> GLU514 A OE1_4017_4018 4.6
@> ARG63 B NH1_5687_5688 <---> ASP129 B OD1_6102_6103 4.6
@> ARG461 B NH1_8778_8779 <---> GLU458 B OE1_8751_8752 4.6
@> ARG444 A NH1_3433_3434 <---> GLU440 A OE1_3397_3398 4.6
@> GLU369 A OE1_2832_2833 <---> HIS368 A NE2_2824 4.6
@> HIS231 B NE2_6962 <---> GLU234 B OE1_6993_6994 4.6
@> LYS165 A NZ_1216 <---> ASP163 A OD1_1197_1198 4.6
@> LYS612 B NZ_9952 <---> ASP562 B OD1_9583_9584 4.7
@> ASP20 A OD1_167_168 <---> LYS71 A NZ_518 4.7
@> GLU212 B OE1_6795_6796 <---> ARG208 B NH1_6764_6765 4.7
@> GLU369 B OE1_8036_8037 <---> HIS368 B NE2_8028 4.8
@> HIS231 A NE2_1758 <---> GLU234 A OE1_1789_1790 4.8
@> GLU168 B OE1_6442_6443 <---> LYS175 B NZ_6501 4.8
@> ARG417 B NH1_8432_8433 <---> GLU382 B OE1_8140_8141 4.9
@> ARG474 B NH1_8890_8891 <---> ASP475 B OD1_8898_8899 4.9
@> ARG215 A NH1_1619_1620 <---> GLU212 A OE1_1591_1592 4.9
@> GLU12 B OE1_5366_5367 <---> ARG90 B NH1_5807_5808 4.9
@> LYS198 B NZ_6680 <---> GLU194 B OE1_6645_6646 5.0
@> Number of detected salt bridges: 64.
@> Calculating repulsive ionic bonding.
@> ASP352 A OD1_2680_2681 <---> ASP349 A OD1_2649_2650 3.3
@> LYS165 A NZ_1216 <---> LYS152 A NZ_1109 3.8
@> ARG203 B NH1_6718_6719 <---> ARG208 B NH1_6764_6765 4.3
@> Number of detected Repulsive Ionic Bonding interactions: 3.
@> Calculating Pi stacking interactions.
@> HIS227 B 6923_6924_6925_6926_6927 <---> HIS231 B 6958_6959_6960_6961_6962 4.1 23.4
@> HIS227 A 1719_1720_1721_1722_1723 <---> HIS231 A 1754_1755_1756_1757_1758 4.1 29.7
@> PHE640 A 4970_4971_4972_4973_4974_4975 <---> PHE487 A 3788_3789_3790_3791_3792_3793 4.3 177.8
@> HIS411 B 8382_8383_8384_8385_8386 <---> TYR176 B 6507_6508_6509_6510_6511_6512 4.5 173.1
@> TRP566 B 9609_9611_9612_9613_9614_9615 <---> PHE229 B 6938_6939_6940_6941_6942_6943 4.5 105.4
@> PHE640 B10174_10175_10176_10177_10178_10179 <---> PHE487 B 8992_8993_8994_8995_8996_8997 4.5 166.4
@> HIS373 B 8063_8064_8065_8066_8067 <---> HIS378 B 8105_8106_8107_8108_8109 4.5 123.3
@> PHE229 A 1734_1735_1736_1737_1738_1739 <---> TRP566 A 4405_4407_4408_4409_4410_4411 4.6 75.3
@> TYR176 A 1303_1304_1305_1306_1307_1308 <---> HIS411 A 3178_3179_3180_3181_3182 4.7 87.1
@> TYR256 B 7170_7171_7172_7173_7174_7175 <---> HIS255 B 7160_7161_7162_7163_7164 4.7 82.3
@> HIS553 B 9520_9521_9522_9523_9524 <---> HIS378 B 8105_8106_8107_8108_8109 4.7 99.2
@> HIS255 A 1956_1957_1958_1959_1960 <---> TYR256 A 1966_1967_1968_1969_1970_1971 4.8 66.4
@> PHE399 A 3072_3073_3074_3075_3076_3077 <---> HIS394 A 3031_3032_3033_3034_3035 4.8 125.6
@> TRP109 B 5954_5956_5957_5958_5959_5960 <---> PHE88 B 5786_5787_5788_5789_5790_5791 4.9 45.5
@> HIS553 A 4316_4317_4318_4319_4320 <---> HIS378 A 2901_2902_2903_2904_2905 4.9 95.9
@> HIS373 A 2859_2860_2861_2862_2863 <---> HIS378 A 2901_2902_2903_2904_2905 5.0 85.4
@> Number of detected Pi stacking interactions: 16.
@> Calculating cation-Pi interactions.
@> PHE399 B 8276_8277_8278_8279_8280_8281 <---> ARG145 B NH1_6242_6243 3.8
@> PHE229 B 6938_6939_6940_6941_6942_6943 <---> LYS214 B NZ_6813 4.4
@> PHE219 B 6857_6858_6859_6860_6861_6862 <---> ARG220 B NH1_6872_6873 4.5
@> HIS376 A 2885_2886_2887_2888_2889 <---> LYS552 A NZ_4310 4.5
@> PHE219 A 1653_1654_1655_1656_1657_1658 <---> ARG220 A NH1_1668_1669 4.6
@> TYR408 B 8355_8356_8357_8358_8359_8360 <---> ARG407 B NH1_8348_8349 4.6
@> PHE399 A 3072_3073_3074_3075_3076_3077 <---> ARG145 A NH1_1038_1039 4.6
@> TYR408 A 3151_3152_3153_3154_3155_3156 <---> ARG407 A NH1_3144_3145 4.6
@> TYR154 B 6324_6325_6326_6327_6328_6329 <---> LYS152 B NZ_6313 4.6
@> PHE344 A 2607_2608_2609_2610_2611_2612 <---> LYS582 A NZ_4530 4.7
@> TYR408 B 8355_8356_8357_8358_8359_8360 <---> LYS180 B NZ_6540 4.7
@> TYR472 B 8862_8863_8864_8865_8866_8867 <---> ARG654 B NH1_10292_10293 4.8
@> HIS160 B 6376_6377_6378_6379_6380 <---> LYS518 B NZ_9256 4.8
@> TYR107 A 727_728_729_730_731_732 <---> ARG90 A NH1_588_589 4.9
@> TYR472 A 3658_3659_3660_3661_3662_3663 <---> ARG654 A NH1_5088_5089 4.9
@> Number of detected cation-pi interactions: 15.
@> Hydrophobic Overlaping Areas are computed.
@> Calculating hydrophobic interactions.
@> ILE433 B CD1_855114s <---> PHE438 B CD1_8583 2.2 42.8
@> MET446 A SD_344914s <---> LEU449 A CD1_3475 2.8 43.8
@> ALA179 B CB_653114s <---> PHE14 B CE2_5382 2.9 48.5
@> ILE421 A CD1_326214s <---> TYR154 A OH_1126 2.9 21.4
@> PHE92 A CE2_61314s <---> VAL69 A CG2_499 3.0 33.3
@> PHE438 A CD1_337914s <---> ILE433 A CG2_3346 3.0 43.4
@> MET478 A SD_371414s <---> ILE460 A CD1_3564 3.0 30.7
@> ILE460 B CG2_876714s <---> VAL465 B CG2_8808 3.0 42.3
@> VAL6 B CG2_532314s <---> LEU94 B CD2_5850 3.1 23.4
@> ARG474 B CG_888614s <---> ILE460 B CD1_8768 3.1 37.5
@> LEU210 B CD1_677814s <---> ILE591 B CG1_9794 3.1 33.1
@> TRP207 B NE1_674914s <---> MET567 B CE_9623 3.1 22.5
@> VAL55 B CG1_562614s <---> LEU36 B CD1_5537 3.1 20.4
@> ILE515 A CG2_402514s <---> TYR541 A OH_4229 3.2 29.9
@> TYR472 B OH_886814s <---> LEU658 B CD2_10322 3.2 31.2
@> ALA123 B CB_605414s <---> TYR495 B CE1_9056 3.2 30.9
@> ARG220 B CG_686814s <---> PHE219 B CE2_6861 3.2 81.3
@> LEU594 A CD1_461114s <---> MET213 A CE_1600 3.2 14.0
@> ILE515 B CG2_922914s <---> TYR541 B OH_9433 3.2 29.6
@> TRP158 B CH2_636314s <---> ILE442 B CD1_8618 3.2 45.7
@> PHE367 A CE2_281314s <---> ILE294 A CG2_2248 3.2 17.1
@> VAL8 A CG2_8414s <---> PHE92 A CD1_610 3.2 28.0
@> PHE184 B CD2_656914s <---> ILE197 A CD1_1467 3.3 29.5
@> TYR664 A CD1_516614s <---> ALA558 A CB_4348 3.3 38.4
@> TRP608 B NE1_991614s <---> ARG220 B CG_6868 3.3 46.3
@> LEU605 B CD1_989314s <---> ALA191 B CB_6621 3.3 16.4
@> TYR472 A OH_366414s <---> LEU658 A CD2_5118 3.3 33.0
@> LEU594 B CD1_981514s <---> MET213 B CE_6804 3.3 16.0
@> ALA188 B CB_660614s <---> LEU609 B CD1_9928 3.3 30.9
@> ALA370 A CB_283814s <---> PHE438 A CD2_3380 3.3 42.4
@> LEU521 A CD1_407414s <---> MET446 A CE_3450 3.3 11.8
@> LEU538 A CD2_420114s <---> ILE492 A CD1_3831 3.3 25.6
@> LEU401 B CD1_829714s <---> PHE487 B CE2_8996 3.3 21.3
@> TYR495 A CE1_385214s <---> ALA123 A CB_850 3.3 28.2
@> VAL24 B CG1_545314s <---> LEU67 B CD1_5718 3.3 11.0
@> PHE104 A CE1_70614s <---> LEU94 A CD1_630 3.3 16.3
@> ILE468 A CG2_362814s <---> TYR471 A CD2_3648 3.3 15.5
@> TRP359 B CZ3_794914s <---> MET574 B CG_9672 3.3 43.2
@> LEU201 B CD1_669914s <---> PHE192 B CE1_6630 3.3 31.1
@> PHE92 B CE2_583214s <---> VAL8 B CG2_5341 3.3 31.8
@> TYR318 A CD1_242314s <---> LEU272 A CD2_2090 3.4 34.9
@> LEU250 B CD2_711814s <---> PHE367 B CZ_8018 3.4 47.0
@> LEU317 A CD1_241514s <---> ILE251 A CD1_1922 3.4 14.3
@> ARG90 A CG_58414s <---> PHE88 A CE2_571 3.4 31.2
@> PHE4 A CD2_4514s <---> LEU57 A CD1_403 3.4 14.5
@> LEU441 A CD1_340514s <---> ILE433 A CD1_3347 3.4 15.4
@> VAL290 A CG2_221914s <---> LEU317 A CD1_2415 3.4 9.6
@> PHE547 A CE1_427514s <---> ALA551 A CB_4301 3.4 31.0
@> PHE219 A CE2_165714s <---> ARG220 A CG_1664 3.4 91.6
@> PHE45 A CZ_31514s <---> LEU38 A CD1_295 3.4 14.4
@> MET148 A CG_106314s <---> TYR149 A CE2_1075 3.4 68.6
@> LEU110 A CD2_76414s <---> TRP87 A CZ3_560 3.4 54.2
@> PHE192 A CZ_142814s <---> LYS196 A CG_1456 3.4 36.2
@> TYR473 A CE2_367414s <---> ALA555 A CB_4330 3.4 13.4
@> PHE384 B CD2_815614s <---> VAL545 B CG1_9461 3.4 38.5
@> TYR496 B CD1_906614s <---> VAL502 B CG2_9114 3.4 32.3
@> ARG417 A CG_322414s <---> ILE421 A CD1_3262 3.4 19.2
@> LEU210 A CD2_157514s <---> MET213 A CE_1600 3.4 42.7
@> LEU456 B CD1_873514s <---> ILE460 B CD1_8768 3.4 39.6
@> VAL263 A CG2_202914s <---> PHE261 A CZ_2015 3.4 35.6
@> VAL597 A CG2_463314s <---> TRP207 A CD2_1544 3.4 51.7
@> LEU355 B CD1_791414s <---> TRP359 B NE1_7945 3.4 38.6
@> TRP511 A CE3_398814s <---> LEU508 A CD1_3962 3.4 36.6
@> LEU605 A CD1_468914s <---> ALA191 A CB_1417 3.4 12.4
@> LEU420 B CD1_845714s <---> VAL426 B CG1_8499 3.4 20.8
@> VAL69 B CG2_573714s <---> PHE92 B CE1_5831 3.4 28.2
@> LEU354 B CD2_790714s <---> TRP232 B CH2_6976 3.4 36.8
@> VAL542 A CG1_423514s <---> LEU401 A CD2_3094 3.4 9.3
@> VAL360 A CG2_275314s <---> ILE331 A CG2_2514 3.5 14.1
@> VAL125 B CG1_606914s <---> TRP127 B CE2_6086 3.5 50.8
@> LYS214 A CD_160714s <---> PHE229 A CZ_1739 3.5 36.1
@> LEU329 B CD2_770614s <---> VAL271 B CG1_7285 3.5 17.7
@> ILE294 B CG2_745214s <---> LEU295 B CD1_7460 3.5 41.1
@> LEU419 B CD1_844914s <---> LYS196 A CD_1457 3.5 34.6
@> LYS518 B CG_925314s <---> TRP151 B CE2_6300 3.5 61.9
@> MET574 A CG_446814s <---> TRP359 A CZ3_2745 3.5 46.6
@> PHE590 B CE2_978714s <---> LEU594 B CD1_9815 3.5 37.1
@> ILE343 B CG2_780414s <---> ALA330 B CB_7711 3.5 3.4
@> PHE547 B CE2_948014s <---> ALA551 B CB_9505 3.5 25.8
..
..
@> Number of detected hydrophobic interactions: 324.
@> Calculating disulfide bonds.
@> Number of detected disulfide bonds: 0.
Select interactions between chains¶
To extract the interactions between protein’s complex, specify selection and selection2 and interaction type:
For hydrogen bonds:
In [6]: interactions.getHydrogenBonds(selection='chain A', selection2='chain B')
[['ARG215', 'NH2_1620', 'A', 'GLU168', 'OE1_6442', 'B', 2.5802, 24.8343],
['ARG215', 'NH1_1619', 'A', 'GLU168', 'OE2_6443', 'B', 2.6778, 28.6548],
['ASP202', 'OD2_1504', 'A', 'GLU418', 'OE2_8442', 'B', 2.744, 31.6383]]
For salt bridges:
In [7]: interactions.getSaltBridges(selection='chain A', selection2='chain B')
[['GLU168', 'OE1_6442_6443', 'B', 'ARG215', 'NH1_1619_1620', 'A', 2.6066],
['ARG208', 'NH1_1560_1561', 'A', 'GLU111', 'OE1_5976_5977', 'B', 4.3468]]
For hydrophobic interactions:
In [8]: interactions.getHydrophobic(selection='chain A', selection2='chain B')
[['PHE184', 'CD2_6569', 'B', 'ILE197', 'CD1_1467', 'A', 3.2502, 29.5284],
['LEU419', 'CD1_8449', 'B', 'LYS196', 'CD_1457', 'A', 3.4645, 34.5683],
['ALA182', 'CB_1349', 'A', 'ILE197', 'CD1_6671', 'B', 3.7348, 34.1782],
['ALA193', 'CB_6637', 'B', 'LEU186', 'CD1_1387', 'A', 4.2965, 20.2503]]
For Pi-stacking interaction:
In [9]: interactions.getPiStacking(selection='chain A', selection2='chain B')
[]
For Pi-cation interactions:
In [10]: interactions.getPiCation(selection='chain A', selection2='chain B')
[]
For repulsive ionic bonding interactions:
In [11]: interactions.getRepulsiveIonicBonding(selection='chain A',
....: selection2='chain B')
....:
[]
Set only interactions between chains¶
With the above functions, we can display particular types of interactions between selected
chains. To set this selection and ignore all intramolecular interactions, we can use
replace option.
In [12]: chain_interactions = interactions.getInteractions(selection='chain A',
....: selection2='chain B', replace=True)
....:
@> New interactions are set
In [13]: chain_interactions
[[['ARG215', 'NH2_1620', 'A', 'GLU168', 'OE1_6442', 'B', 2.5802, 24.8343],
['ARG215', 'NH1_1619', 'A', 'GLU168', 'OE2_6443', 'B', 2.6778, 28.6548],
['ASP202', 'OD2_1504', 'A', 'GLU418', 'OE2_8442', 'B', 2.744, 31.6383]],
[['ARG215', 'NH1_1619_1620', 'A', 'GLU168', 'OE1_6442_6443', 'B', 2.6066],
['ARG208', 'NH1_1560_1561', 'A', 'GLU111', 'OE1_5976_5977', 'B', 4.3468]],
[],
[],
[],
[['ILE197', 'CD1_1467', 'A', 'PHE184', 'CD2_6569', 'B', 3.2502, 29.5284],
['LEU419', 'CD1_8449', 'B', 'LYS196', 'CD_1457', 'A', 3.4645, 34.5683],
['ILE197', 'CD1_6671', 'B', 'ALA182', 'CB_1349', 'A', 3.7348, 34.1782],
['LEU186', 'CD1_1387', 'A', 'ALA193', 'CB_6637', 'B', 4.2965, 20.2503]],
[]]
Now, when new interactions are set, we can use the functions that we introduced before:
Interaction matrix:
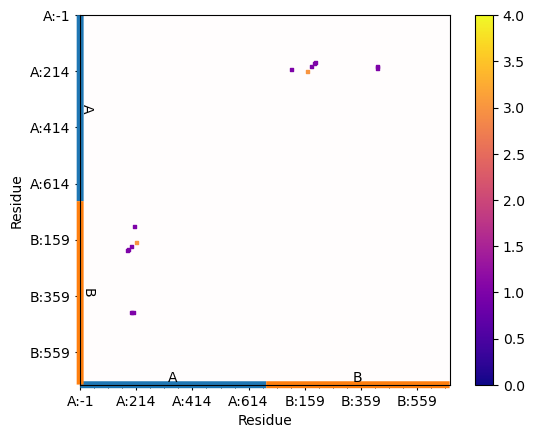
In [14]: matrix = interactions.buildInteractionMatrix()
@> Calculating interaction matrix
In [15]: import matplotlib.pylab as plt
In [16]: showAtomicMatrix(matrix, atoms=atoms.ca, cmap='plasma', markersize=5)
In [17]: plt.xlabel('Residue')
In [18]: plt.ylabel('Residue')
In [19]: plt.clim([0,np.max(matrix)+1])
Interaction matrix displayed with energies:
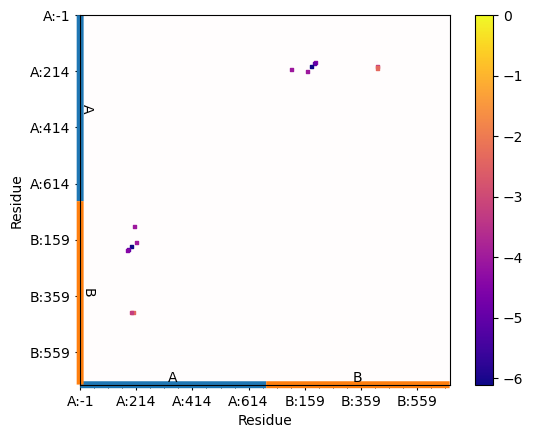
In [20]: matrix_en = interactions.buildInteractionMatrixEnergy()
@> Calculating interaction energies matrix with type IB_solv
In [21]: import matplotlib.pylab as plt
In [22]: showAtomicMatrix(matrix_en, atoms=atoms.ca, cmap='plasma',
....: markersize=5)
....:
In [23]: plt.xlabel('Residue')
In [24]: plt.ylabel('Residue')
In [25]: plt.clim([np.min(matrix_en),0])
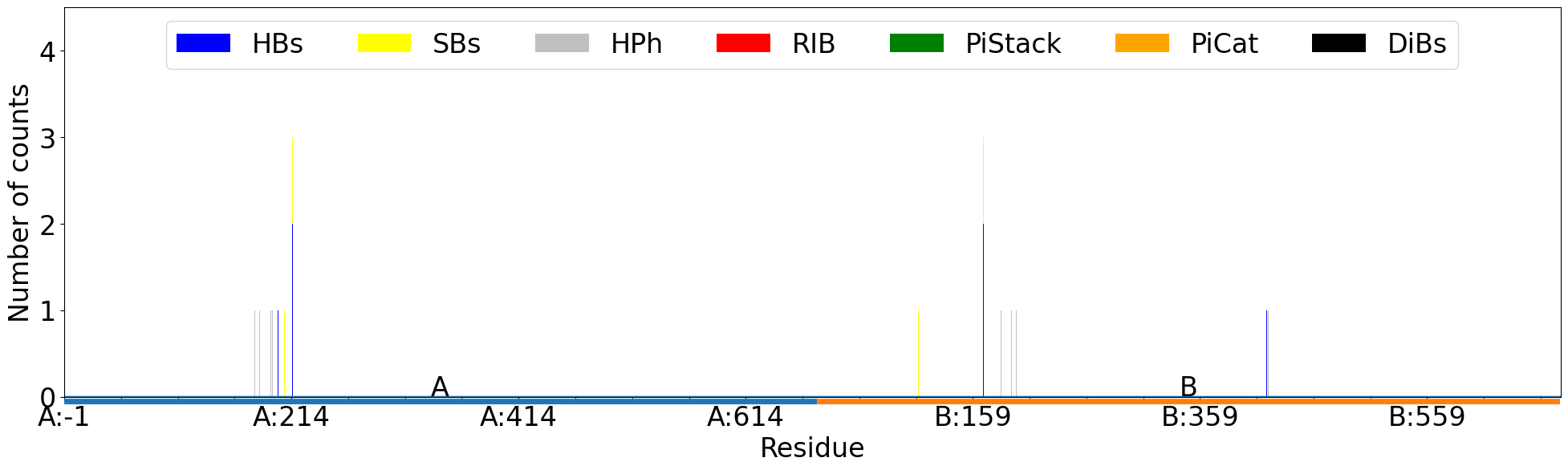
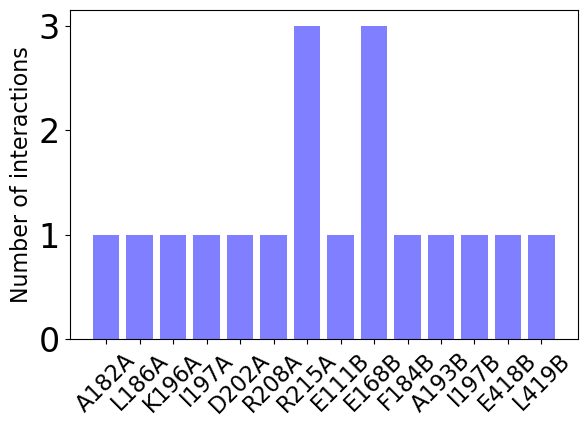
A bar plot with information about interaction types per residue between the two chains:
In [26]: interactions.showCumulativeInteractionTypes()
@> Calculating interaction matrix
@> Calculating interaction matrix
@> Calculating interaction matrix
@> Calculating interaction matrix
@> Calculating interaction matrix
@> Calculating interaction matrix
@> Calculating interaction matrix
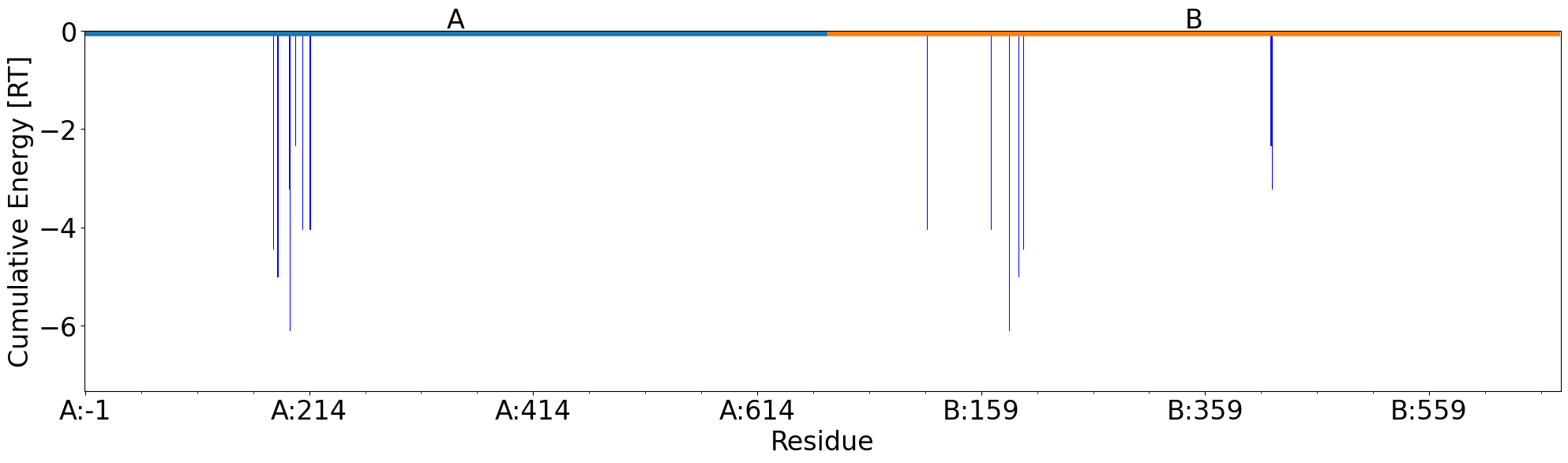
A bar plot with information about interaction types per residue between the two chains displayed by energies instead of the interactions number:
In [27]: interactions.showCumulativeInteractionTypes(energy=True)
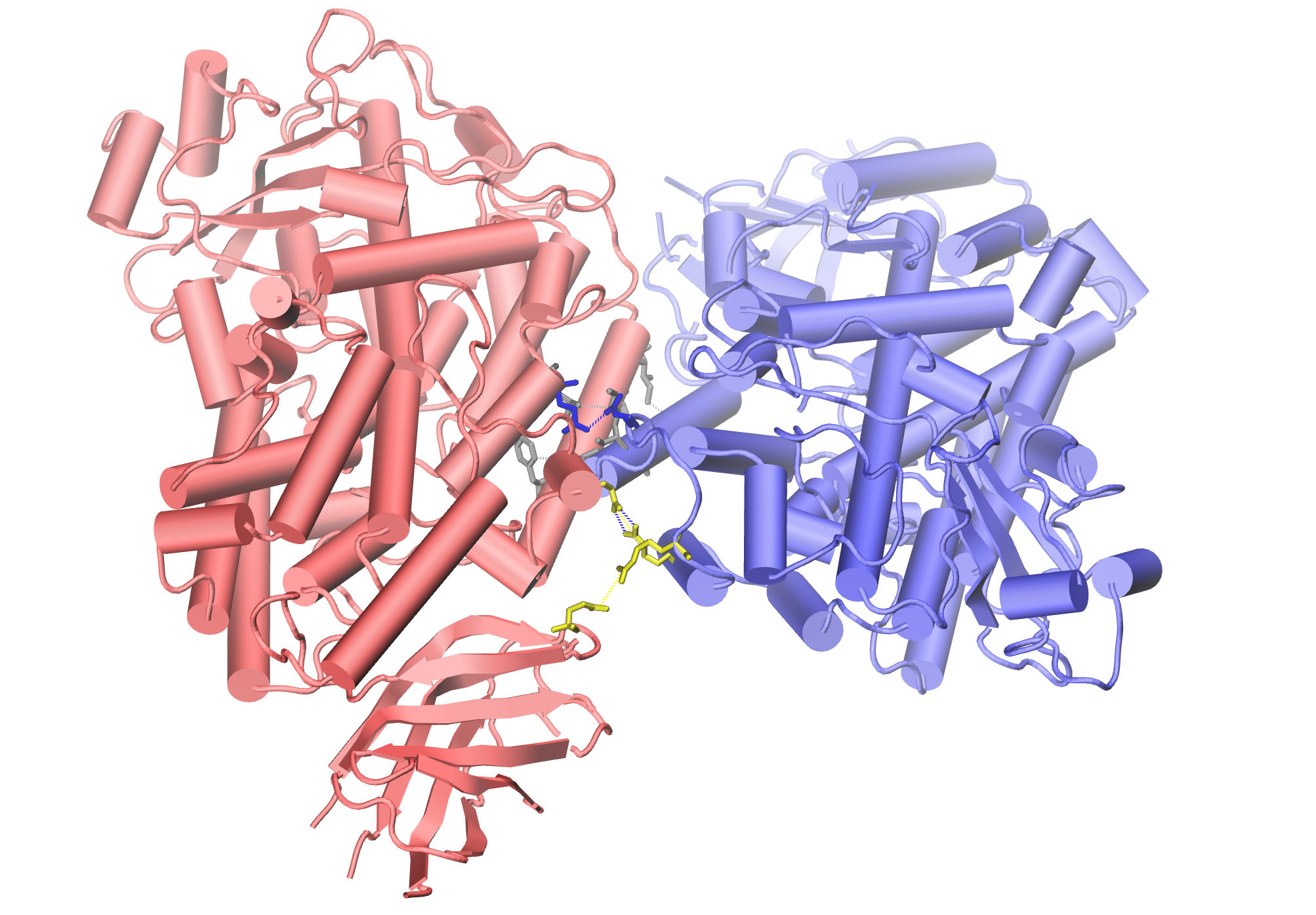
The results with the higest number of possible contacts can be saved in PDB file. They will be restored in Occupancy column and display in VMD.
In [28]: interactions.saveInteractionsPDB(filename='7laf_meanMatrix_chAB_both.pdb')
@> PDB file saved.
Also, with enegies instead of the number of interactions:
In [29]: interactions.saveInteractionsPDB(filename='7laf_meanMatrix_chAB_both_en.pdb', energy=True)
@> PDB file saved.
We can also save all the interactions into the TCL scripts to visualize
them in VMD.
In [30]: showProteinInteractions_VMD(atoms, interactions.getHydrogenBonds(),
....: color='blue', filename='7laf_HBs_chAB.tcl')
....:
In [31]: showProteinInteractions_VMD(atoms, interactions.getSaltBridges(),
....: color='yellow',filename='7laf_SBs_chAB.tcl')
....:
In [32]: showProteinInteractions_VMD(atoms, interactions.getRepulsiveIonicBonding(),
....: color='red',filename='7laf_RIB_chAB.tcl')
....:
In [33]: showProteinInteractions_VMD(atoms, interactions.getPiStacking(),
....: color='green',filename='7laf_PiStacking_chAB.tcl')
....:
In [34]: showProteinInteractions_VMD(atoms, interactions.getPiCation(),
....: color='orange',filename='7laf_PiCation_chAB.tcl')
....:
In [35]: showProteinInteractions_VMD(atoms, interactions.getHydrophobic(),
....: color='silver',filename='7laf_HPh_chAB.tcl')
....:
In [36]: showProteinInteractions_VMD(atoms, interactions.getDisulfideBonds(),
....: color='black',filename='7laf_DiBs_chAB.tcl')
....:
@> TCL file saved
@> TCL file saved
@> Lack of results
@> TCL file saved
@> Lack of results
@> TCL file saved
@> Lack of results
@> TCL file saved
@> TCL file saved
@> Lack of results
@> TCL file saved
After uploading TCL scripts to VMD, as it was explained before, we can
obtain such a view:
Except for visualization, we can also get access to the most frequent interactors, i.e., residues that can form the biggest number of possible interactions.
In [37]: interactions.getFrequentInteractors()
@> GLU168B <---> hb:ARG215A hb:ARG215A sb:ARG215A
@> ARG215A <---> hb:GLU168B hb:GLU168B sb:GLU168B
@>
Legend: hb-hydrogen bond, sb-salt bridge, rb-repulsive ionic bond,
ps-Pi stacking interaction, pc-Cation-Pi interaction, hp-hydrophobic
interaction, dibs-disulfide bonds
To have access to interactors that are having smaller number of
interactions, we can modify contacts_min parameter.
In [38]: interactions.getFrequentInteractors(contacts_min=1)
@> ALA182A <---> hp:ILE197B
@> LYS196A <---> hp:LEU419B
@> GLU111B <---> sb:ARG208A
@> GLU168B <---> hb:ARG215A hb:ARG215A sb:ARG215A
@> PHE184B <---> hp:ILE197A
@> ALA193B <---> hp:LEU186A
@> GLU418B <---> hb:ASP202A
@> ILE197B <---> hp:ALA182A
@> LEU419B <---> hp:LYS196A
@> ARG208A <---> sb:GLU111B
@> ARG215A <---> hb:GLU168B hb:GLU168B sb:GLU168B
@> ILE197A <---> hp:PHE184B
@> LEU186A <---> hp:ALA193B
@> ASP202A <---> hb:GLU418B
@>
Legend: hb-hydrogen bond, sb-salt bridge, rb-repulsive ionic bond, ps-Pi stacking interaction,
pc-Cation-Pi interaction, hp-hydrophobic interaction, dibs-disulfide bonds
We can also diplay them as a bar plot:
In [39]: interactions.showFrequentInteractors(cutoff=1)
To have access to information about the type of possible interactions and
residue partner, we can use the getInteractors() function and define
residue by its three letter code and chain ID.
In [40]: interactions.getInteractors('ARG215A')
@> hb:ARG215A-GLU168B
@> hb:ARG215A-GLU168B
@> sb:ARG215A-GLU168B
Extract chain-chain interactions in ensemble or trajectory analysis¶
To extract the intermolecular interactions between two chains in a
trajectory or PDB ensemble, we should follow the corresponding tutorial and
include selections (selection and selection2) and the replace
parameter set to True as follows:
In [41]: interactionsTrajectory.getInteractions(selection='chain A',
....: selection2='chain B', replace=True)
....:
Once we use replace = True, the selection will be replaced by
chain-chain interactions or any other interaction selected by selecting
option. Be aware that once replace is used, you can not return back to all
interactions.