Analyze Conformations¶
First, necessary imports:
In [1]: from prody import *
In [2]: from numpy import *
In [3]: from pylab import *
In [4]: ion()
In [5]: import os, glob
Parse conformations¶
Now, let’s read initial and refined conformations:
In [6]: initial = AtomGroup('p38 initial')
In [7]: refined = AtomGroup('p38 refined')
In [8]: for pdb in glob.glob('p38_ensemble/*pdb'):
...: fn = os.path.splitext(os.path.split(pdb)[1])[0]
...: opt = os.path.join('p38_optimize', fn + '.coor')
...: parsePDB(pdb, ag=initial)
...: parsePDB(opt, ag=refined)
...:
---------------------------------------------------------------------------
MMCIFParseError Traceback (most recent call last)
<ipython-input-8-87036b051e71> in <module>()
3 opt = os.path.join('p38_optimize', fn + '.coor')
4 parsePDB(pdb, ag=initial)
----> 5 parsePDB(opt, ag=refined)
6
/home/exx/ProDy-website/ProDy/prody/proteins/pdbfile.pyc in parsePDB(*pdb, **kwargs)
125
126 if n_pdb == 1:
--> 127 return _parsePDB(pdb[0], **kwargs)
128 else:
129 results = []
/home/exx/ProDy-website/ProDy/prody/proteins/pdbfile.pyc in _parsePDB(pdb, **kwargs)
244 try:
245 LOGGER.warn("Trying to parse as mmCIF file instead")
--> 246 return parseMMCIF(pdb, **kwargs)
247 except KeyError:
248 try:
/home/exx/ProDy-website/ProDy/prody/proteins/ciffile.pyc in parseMMCIF(pdb, **kwargs)
130 kwargs['title'] = title
131 cif = openFile(pdb, 'rt')
--> 132 result = parseMMCIFStream(cif, chain=chain, segment=segment, **kwargs)
133 cif.close()
134 if unite_chains:
/home/exx/ProDy-website/ProDy/prody/proteins/ciffile.pyc in parseMMCIFStream(stream, **kwargs)
237
238 _parseMMCIFLines(ag, lines, model, chain, subset, altloc,
--> 239 segment, unite_chains, report)
240
241 if ag.numAtoms() > 0:
/home/exx/ProDy-website/ProDy/prody/proteins/ciffile.pyc in _parseMMCIFLines(atomgroup, lines, model, chain, subset, altloc_torf, segment, unite_chains, report)
337 stop = i
338 else:
--> 339 raise MMCIFParseError('mmCIF file contained no atoms.')
340
341 i += 1
MMCIFParseError: mmCIF file contained no atoms.
In [9]: initial
Out[9]: <AtomGroup: p38 initial (5658 atoms)>
In [10]: refined
Out[10]: <AtomGroup: p38 refined (0 atoms; no coordinates)>
Calculate RMSD change¶
We can plot RMSD change after refinement as follows:
In [11]: rmsd_ca = []
In [12]: rmsd_all = []
In [13]: initial_ca = initial.ca
In [14]: refined_ca = refined.ca
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
<ipython-input-14-40de9febca08> in <module>()
----> 1 refined_ca = refined.ca
/home/exx/ProDy-website/ProDy/prody/atomic/atomic.pyc in __getattribute__(self, name)
98 dummies = self.numDummies()
99 except AttributeError:
--> 100 indices = self._getSubset(name)
101 if len(indices):
102 return Selection(ag, indices, selstr,
/home/exx/ProDy-website/ProDy/prody/atomic/atomgroup.pyc in _getSubset(self, label)
1086 return self._subsets[label]
1087 except KeyError:
-> 1088 flgs = self._getFlags(label)
1089 try:
1090 return self._subsets[label]
/home/exx/ProDy-website/ProDy/prody/atomic/atomgroup.pyc in _getFlags(self, label)
1028 except KeyError:
1029 try:
-> 1030 return FLAG_PLANTERS[label](self, label)
1031 except KeyError:
1032 pass
/home/exx/ProDy-website/ProDy/prody/atomic/flags.pyc in setCalpha(ag, label)
790
791 flags = ag._getNames() == 'CA'
--> 792 indices = flags.nonzero()[0]
793 if len(indices):
794 torf = array([rn in AMINOACIDS for rn in ag._getResnames()[indices]])
AttributeError: 'bool' object has no attribute 'nonzero'
In [15]: for i in range(initial.numCoordsets()):
....: initial.setACSIndex(i)
....: refined.setACSIndex(i)
....: initial_ca.setACSIndex(i)
....: refined_ca.setACSIndex(i)
....: rmsd_ca.append(calcRMSD(initial_ca, refined_ca))
....: rmsd_all.append(calcRMSD(initial, refined))
....:
---------------------------------------------------------------------------
IndexError Traceback (most recent call last)
<ipython-input-15-71aab0f1ba84> in <module>()
1 for i in range(initial.numCoordsets()):
2 initial.setACSIndex(i)
----> 3 refined.setACSIndex(i)
4 initial_ca.setACSIndex(i)
5 refined_ca.setACSIndex(i)
/home/exx/ProDy-website/ProDy/prody/atomic/atomgroup.pyc in setACSIndex(self, index)
858 raise TypeError('index must be an integer')
859 if n_csets <= index or n_csets < abs(index):
--> 860 raise IndexError('coordinate set index is out of range')
861 if index < 0:
862 index += n_csets
IndexError: coordinate set index is out of range
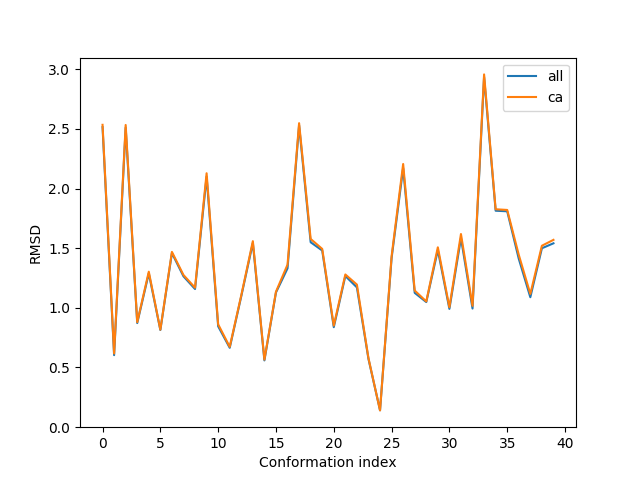
In [16]: plot(rmsd_all, label='all');
In [17]: plot(rmsd_ca, label='ca');
In [18]: xlabel('Conformation index');
In [19]: ylabel('RMSD');
In [20]: legend();
Select a diverse set¶
To select a diverse set of refined conformations, let’s calculate average RMSD for each conformation to all others:
In [21]: rmsd_mean = []
In [22]: for i in range(refined.numCoordsets()):
....: refined.setACSIndex(i)
....: alignCoordsets(refined)
....: rmsd = calcRMSD(refined)
....: rmsd_mean.append(rmsd.sum() / (len(rmsd) - 1))
....:
In [23]: bar(arange(1, len(rmsd_mean) + 1), rmsd_mean);
In [24]: xlabel('Conformation index');
In [25]: ylabel('Mean RMSD');
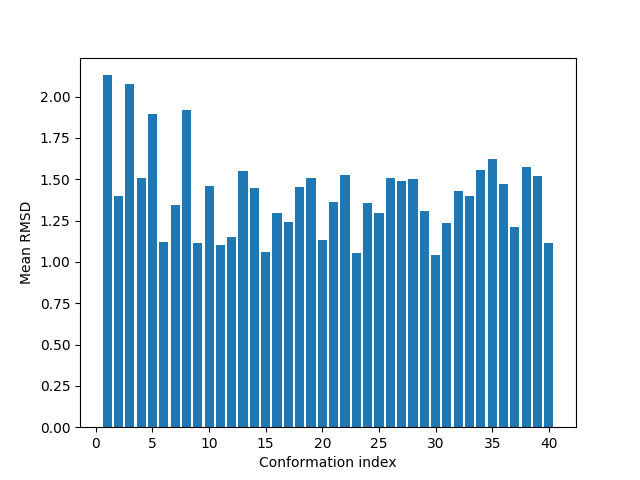
Let’s select conformations that are 1.2 Å away from other on average:
In [26]: selected = (array(rmsd_mean) >= 1.2).nonzero()[0]
In [27]: selected
Out[27]: array([], dtype=int64)
In [28]: selection = refined[selected]
---------------------------------------------------------------------------
IndexError Traceback (most recent call last)
<ipython-input-28-691a562240aa> in <module>()
----> 1 selection = refined[selected]
/home/exx/ProDy-website/ProDy/prody/atomic/atomgroup.pyc in __getitem__(self, index)
208 elif isinstance(index, (list, np.ndarray)):
209 unique = np.unique(index)
--> 210 if unique[0] < 0 or unique[-1] >= self._n_atoms:
211 raise IndexError('index out of range')
212 return Selection(self, unique, 'index ' + rangeString(index),
IndexError: index 0 is out of bounds for axis 0 with size 0
In [29]: selection
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
<ipython-input-29-ae784f1d4d09> in <module>()
----> 1 selection
NameError: name 'selection' is not defined

