|
 |
|
Druggability Simulations and X-ray Crystallography Reveal a Ligand-binding Site in the GluA3 AMPA Receptor N-terminal Domain |
Jiyoung Lee, James Krieger, Beatriz Herguedas, Javier Garcia-Nafria, Anindita Dutta, Saher A. Shaikh, Ingo H. Greger, and Ivet Bahar
|
| |
Ionotropic glutamate receptors (iGluRs) mediate the vast majority of excitatory neurotransmission in the brain. Their dysfunction is implicated in several neurological disorders, rendering iGluRs potential drug targets. Here, we performed a systematic analysis of the druggability of two major iGluR subfamilies, using molecular dynamics simulations in the presence of drug-like molecules. We demonstrate the applicability of druggability simulations by faithfully identifying known agonist and modulator sites on AMPA receptors (AMPARs) (see Fig 1A) and NMDA receptors. Simulations produced the expected allosteric changes of the AMPAR ligand-binding domain in response to agonist. We also identified a novel ligand-binding site specific to the GluA3 AMPAR N-terminal domain (NTD), resulting from its unique conformational flexibility that we explored further with new crystal structures trapped in vastly different states (see Fig 1B). In addition to providing novel insights into iGluR NTD dynamics, our approach identifies druggable sites and permits the determination of pharmacophoric features towards novel iGluR modulators (see Fig 1C).
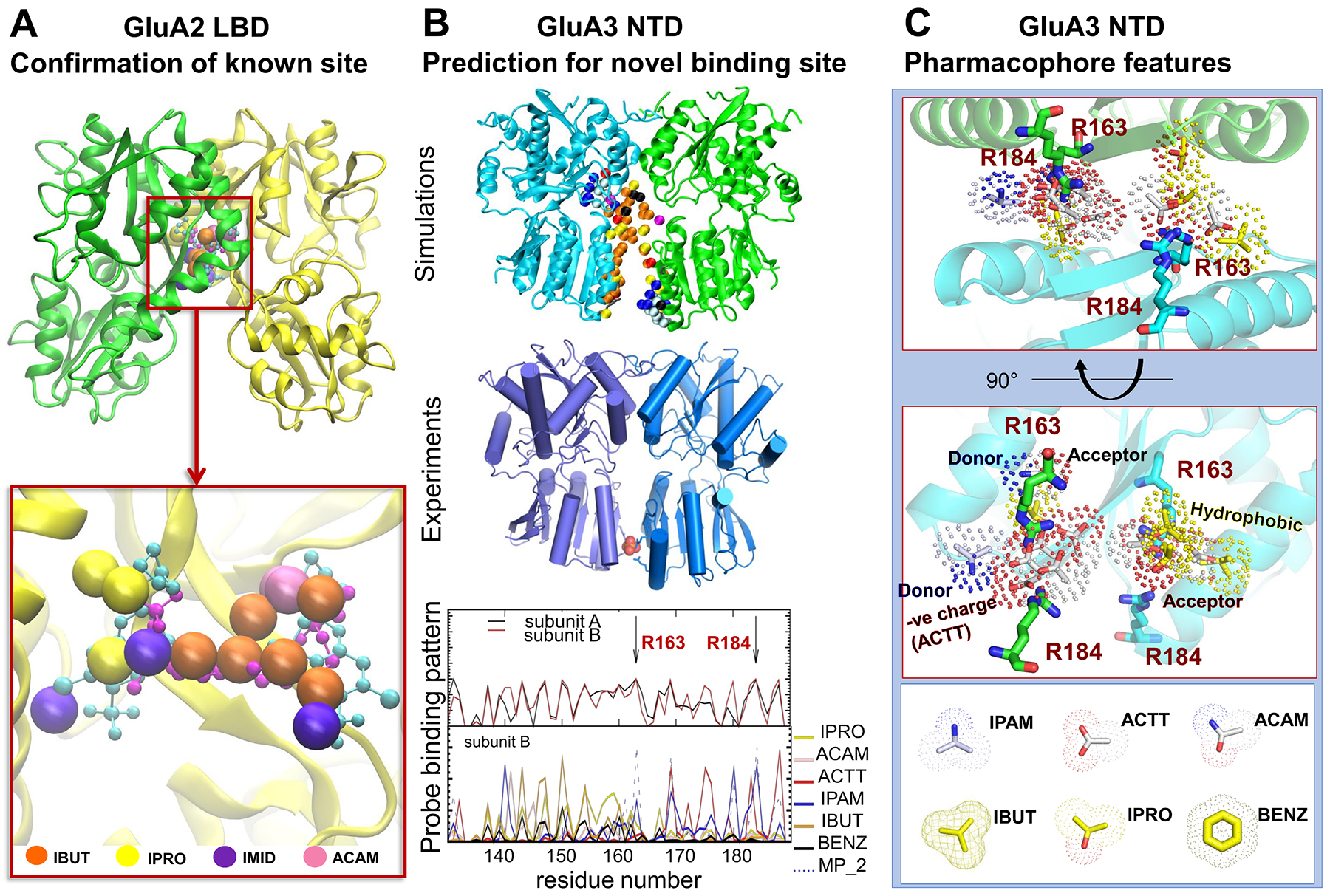
Figure 1. Druggability Simulations, X-Ray Crystallography, and Pharmacophore model for AMPA receptor (A) Druggability MD detecting a known ligand binding site of GluA2 LBD: Large balls are hot spots by probe molecules and their colors are different probes. They closely overlap with the experimentally observed positions of the allosteric modulators cyclothiazide (cyan balls/stick; from PDB ID: 1LBC) and (R,R)-2b (magenta balls/sticks; PDB ID: 4U5B). (B) Novel ligand binding site in GluA3 NTD. (top) Druggability MD probes are shown as spheres near the LL interfaces of the GluA3 NTD dimer. (middle) New crystal structure of the GluA3 NTD reveal new dimeric state, which is similar to the open dimer. (bottom) Number of contacts between probe molecules and residues at the LL interface are observed in the MD simulations. (C) Pharmacophore model in GluA3 NTD LL interface.
|
| |
Related publication:
Lee JY, Krieger J, Herguedas B, García-Nafría J, Dutta A, Shaikh SA, Greger IH, Bahar I. (2019) Druggability Simulations and X-Ray Crystallography Reveal a Ligand-Binding Site in the GluA3 AMPA Receptor N-Terminal Domain Structure 27: 241-252. PMID: 30528594 PMCID: PMC6365164 [Available on 2020-02-05]
Download paper Supplemental Information
Structure: Volume 27, Issue 2
Jiyoung Lee, James Krieger, Anindita Dutta, and Ivet Bahar are affiliated with the Department of Computational and Systems Biology, University of Pittsburgh School of Medicine, 3501 Fifth Ave, Suite 3064 BST3, Pittsburgh, PA 15213 USA
Beatriz Herguedas, Javier García-Nafría, Saher A. Shaikh, and Ingo H. Greger are affiliated with The MRC Laboratory of Molecular Biology (LMB), Neurobiology Division, AMPA receptor biogenesis, structure and function group, Cambridge University, Francis Crick Ave., Cambridge, CB2 0QH, UK
|
|
|
|
|
|
|

